

高等学校化学学报 ›› 2021, Vol. 42 ›› Issue (11): 3468.doi: 10.7503/cjcu20210425
收稿日期:2021-06-22
出版日期:2021-11-10
发布日期:2021-11-10
通讯作者:
陈岚岚,胡善文
E-mail:llchen@fzu.edu.cn;shanwenhu@fjmu.edu.cn
基金资助:
ZHANG Xiaorong1, CHEN Lanlan2( ), HU Shanwen1(
), HU Shanwen1( )
)
Received:2021-06-22
Online:2021-11-10
Published:2021-11-10
Contact:
CHEN Lanlan,HU Shanwen
E-mail:llchen@fzu.edu.cn;shanwenhu@fjmu.edu.cn
Supported by:摘要:
以抗体-抗原免疫识别、 核酸碱基互补配对识别以及核酸适体-配体识别这3种分子识别方式分类, 综述了近几年基于分子识别的细菌检测研究工作进展, 总结了细菌检测相关研究存在的一些挑战, 并展望了该领域的发展前景.
中图分类号:
TrendMD:
张晓荣, 陈岚岚, 胡善文. 基于分子识别的细菌检测研究进展. 高等学校化学学报, 2021, 42(11): 3468.
ZHANG Xiaorong, CHEN Lanlan, HU Shanwen. Advances in Bacteria Biosensing Based on Molecular Recognition. Chem. J. Chinese Universities, 2021, 42(11): 3468.
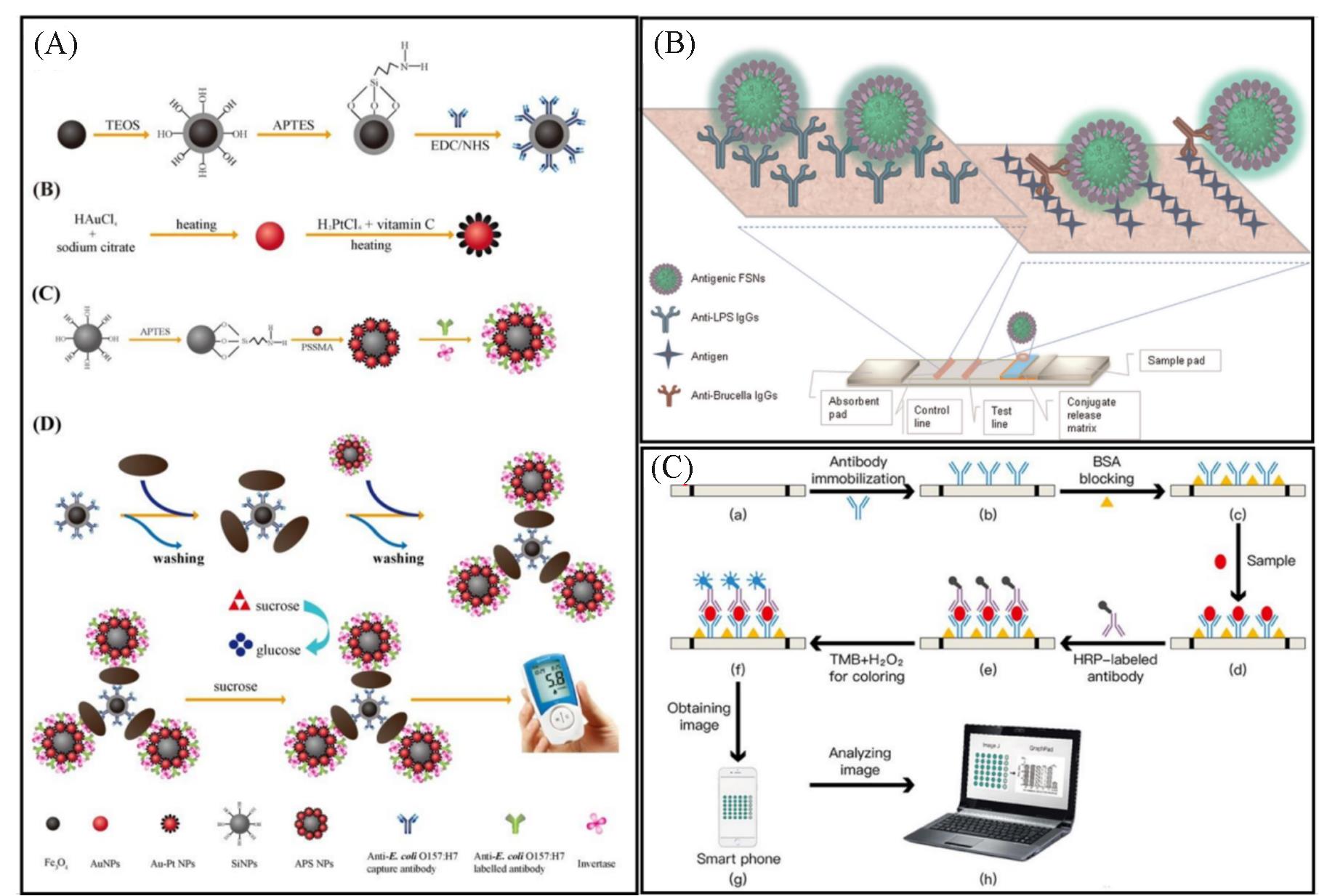
Fig.1 Molecular recognition methods of bacteria based on immune(A) Schematic illustration of recognition process detection of E. coli using Ab?Fe3O4@SiO2 nanoparticles in sandwich?type immunoassay[21]; (B) detailed procedure of immunochromatographic strip test for detection of brucellosis[22]; (C) schematic illustration for E. coli detection by using immunological recognition based paper?ELISA[24].(A) Copyright 2018, Elsevier; (B) Copyright 2015, Elsevier; (C) Copyright 2020, Royal Society of Chemistry.
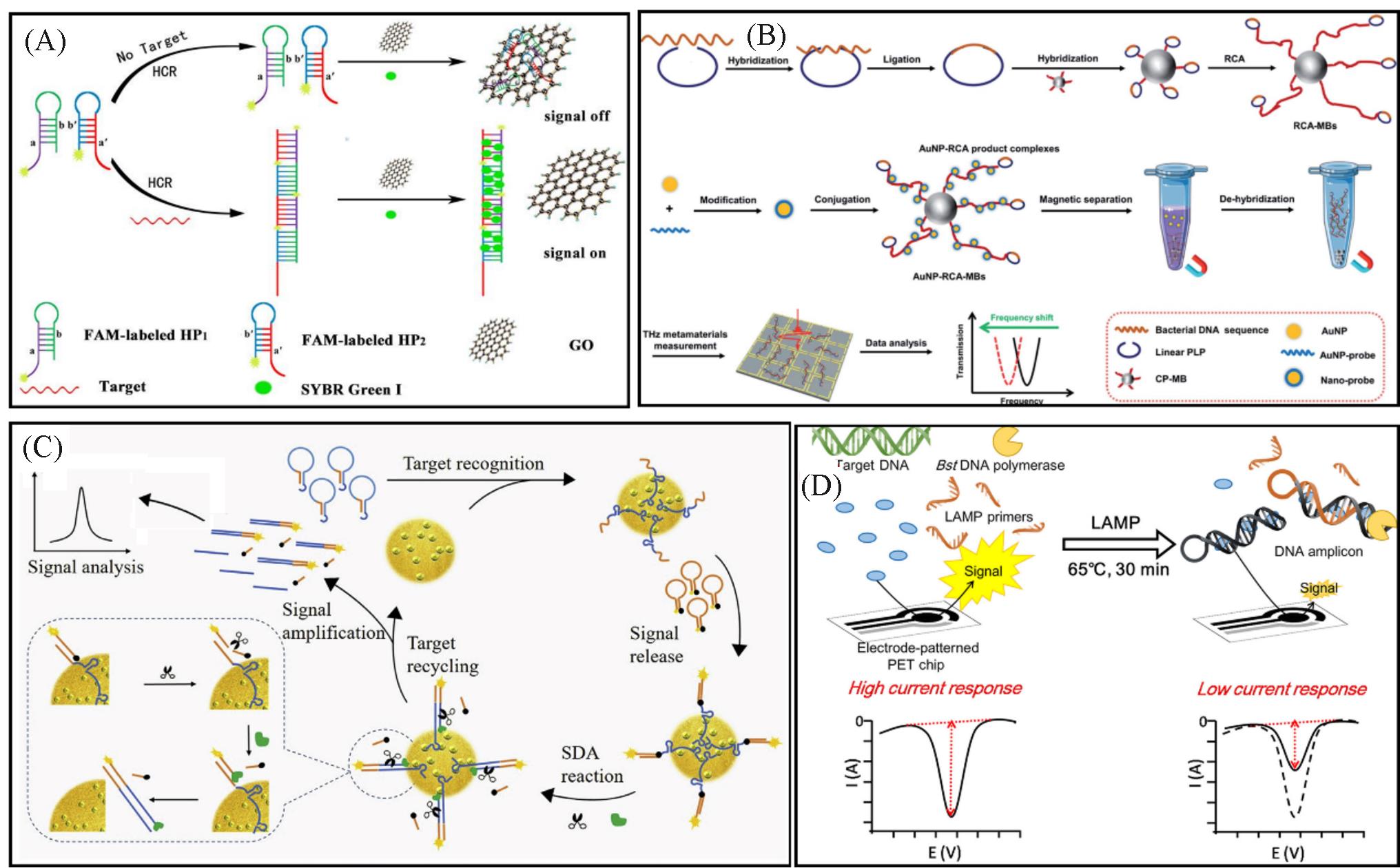
Fig.2 Detection of bacteria based on nucleic acid base pairing recognition methods(A) Schematic illustration of a sensitive and selective enzyme?free Staphylococcus aureus(S. aureus) detection method combined with the characteristic of graphene oxide(GO) and the HCR amplification[36]; (B) principle of the THz metamaterial biosensor by using RCA for bacterial DNA detection[40]; (C) schematic of the fluorescent detection of S. aureus based on SDR[43]; (D) schematic illustration of the detection mechanism for E. coli by using real?time electrochemical LAMP[48]. (A) Copyright 2019, Elsevier; (B) Copyright 2020, Royal Society of Chemistry; (C) Copyright 2019, Elsevier; (D) Copyright 2018, Wiley.
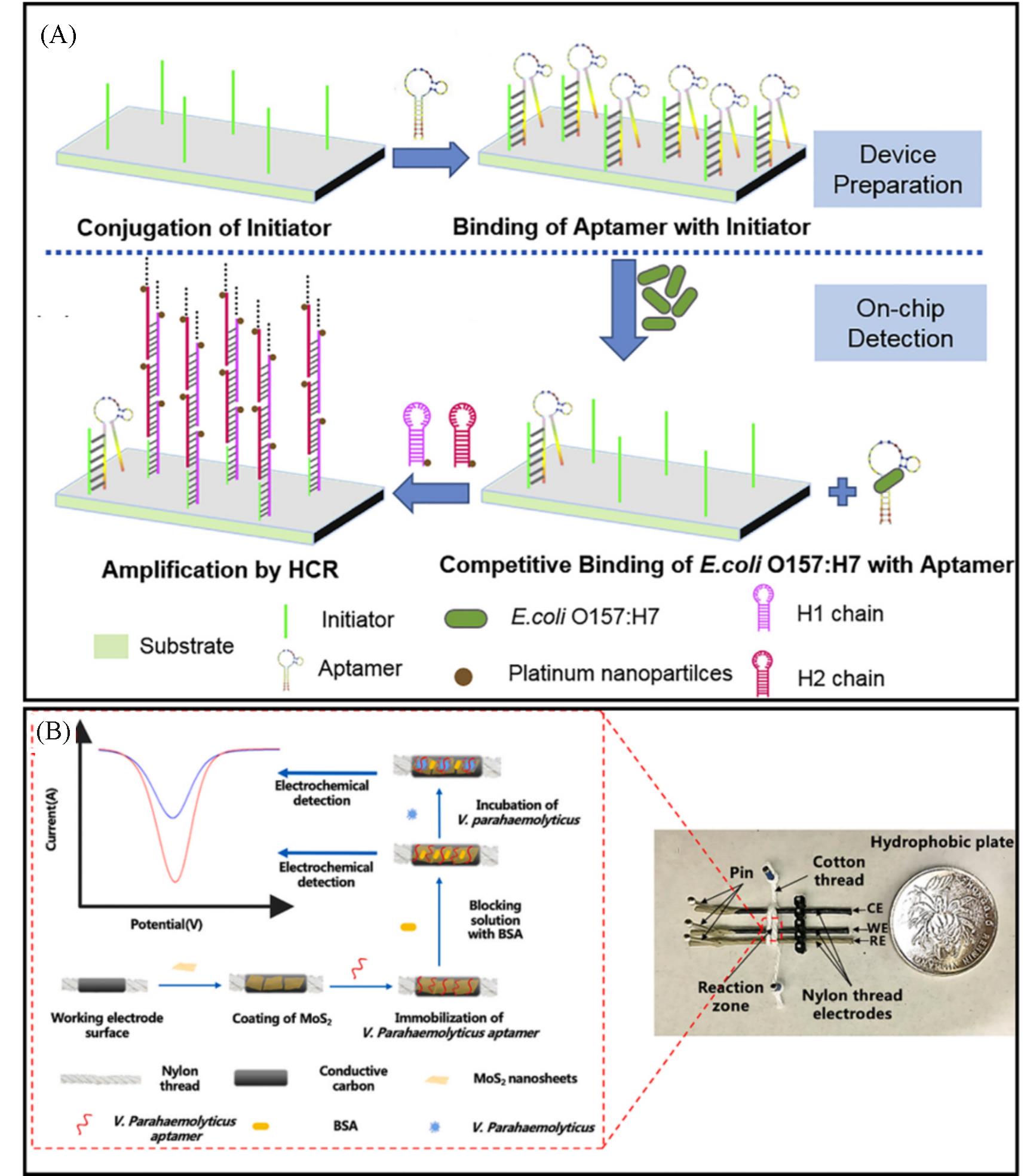
Fig.3 Molecular recognition for bacteria detection based on aptamer binding(A) Schematic representation for E. coli by integrating aptamer recognition, HCR amplification and naked?eye based detection[85]; (B) schematic illustration of the principle of thread?based microfluidic aptasensor for Vibrio parahaemolyticus(V. parahaemolyticus) detection[86].(A) Copyright 2020, Elsevier; (B) Copyright 2021, Elsevier.
| 1 | Kenny D., Balskus E., Chem. Soc. Rev., 2018, 47, 1705―1729 |
| 2 | Rook G., Bäckhed F., Levin B., McFall⁃Ngai M., McLean A., Lancet, 2017, 390, 521―530 |
| 3 | Chen Y., Fischbach M., Belkaid Y., Nature, 2018, 553, 427―436 |
| 4 | Sonnenburg J., Sonnenburg E., Science,2019, 366, eaaw9255 |
| 5 | Fernandez R. E., Rohani A., Farmehini V., Swami N. S., Anal. Chim. Acta, 2017, 966, 11―33 |
| 6 | Li W. S., Wu G. R., Zhang X. J., Yue A. Q., Du W. J., Zhao J. Z., Liu D. B., Chem. J. Chinese Universities, 2020, 41(5), 872―883 (李文帅, 武国瑞, 张茜菁, 岳爱琴, 杜维俊, 赵晋忠, 刘定斌. 高等学校化学学报, 2020, 41(5), 872―883) |
| 7 | Furst A., Francis M., Chem. Rev., 2019, 119, 700―726 |
| 8 | Gentile C., Weir T., Science,2018, 362, 776―780 |
| 9 | Atarashi K., Suda W., Luo C., Kawaguchi T., Motoo I., Narushima S., Kiguchi Y., Yasuma K., Watanabe E., Tanoue T., Thaiss C., Sato M., Toyooka K., Said H., Yamagami H., Rice S., Gevers D., Johnson R., Segre J., Chen K., Kolls J., Elinav E., Morita H., Xavier R., Hattori M., Honda K., Science,2017, 358, 359―365 |
| 10 | Sepich⁃Poore G., Zitvogel L., Straussman R., Hasty J., Wargo J., Knight R., Science,2021, 371, eabc4552 |
| 11 | Lin L., Du Y., Song J., Wang W., Yang C., Acc. Chem. Res., 2021, 54, 2076―2087 |
| 12 | Kim K., Park S., Park C., Seo S., Lee J., Kim J., Lee S., Lee S., Kim J., Ryu C., Yong D., Yoon H., Song H., Lee S., Kwon O., Biosens. Bioelectron., 2020, 167, 112514 |
| 13 | Liang T., Leung L. M., Opene B., Fondrie W. E., Lee Y. I., Chandler C. E., Yoon S. H., Doi Y., Ernst R. K., Goodlett D. R., Anal. Chem., 2019, 91, 1286―1294 |
| 14 | Zhao Q., Lu D., Zhang G. Y., Zhang D., Shi X. B., Talanta, 2021, 223, 121722 |
| 15 | Shen J., Li Y., Gu H., Xia F., Zuo X., Chem. Rev., 2014, 114, 7631―7677 |
| 16 | Wu L., Li G. H., Xu X., Zhu L., Huang R. M., Chen X. Q., Trac⁃Trends Anal. Chem., 2019, 113, 140―156 |
| 17 | Nouri A., Ahari H., Shahbazzadeh D., Int. J. Biol. Macromol., 2018, 107, 1732―1737 |
| 18 | Deng J., Zhao S., Liu Y., Liu C., Sun J., ACS Appl. Bio. Mater., 2020, 4, 3863―3879 |
| 19 | Zhang Y., Shi S. Y., Xing J. J., Tan W. Q., Zhang C. G., Zhang L., Yuan H., Zhang M. M., Qiao J. J., RSC Adv., 2019, 9, 33589―33595 |
| 20 | Bonnet R., Farre C., Valera L., Vossier L., Leon F., Dagland T., Pouzet A., Jaffrezic⁃Renault N., Fareh J., Fournier⁃Wirth C., Chaix C., Analyst, 2018, 143, 2293―2303 |
| 21 | Ye L., Zhao G., Dou W., Talanta, 2018, 182, 354―362 |
| 22 | Vyas S., Jadhav S., Majee S., Shastri J., Patravale V., Biosens. Bioelectron., 2015, 70, 254―260 |
| 23 | Pang B., Zhao C., Li L., Song X. L., Xu K., Wang J., Liu Y. S., Fu K. Y., Bao H., Song D. D., Meng X. J., Qu X. F., Zhang Z. P., Li J., Anal. Biochem., 2018, 542, 58―62 |
| 24 | Zhao Y. A., Zeng D. X., Yan C., Chen W., Ren J. L., Jiang Y., Jiang L. Y., Xue F., Ji D. J., Tang F., Zhou M. Q., Dai J. J., Analyst, 2020, 145, 3106―3115 |
| 25 | Deng M. Y., Li M., Mao X. H., Li F., Zuo X. L., Chem. Res. Chinese Universities, 2020, 36(2), 185―193 |
| 26 | Huang L., Sun D. W., Wu Z., Pu H., Wei Q., Anal. Chim. Acta, 2021, 1167, 338570 |
| 27 | Zhang Y. N., Yang L. L., Tu J. W., Cui R., Pang D. W., Chem. J. Chinese Universities, 2018, 39(6), 1158―1163(张亚楠, 杨玲玲, 涂家薇, 崔然, 庞代文. 高等学校化学学报, 2018, 39(6), 1158―1163) |
| 28 | Xu S., Int. J. Electrochem. Sci.,2017, 3443―3458 |
| 29 | Melaine F., Saad M., Faucher S., Tabrizian M., Anal. Chem., 2017, 89, 7802―7807 |
| 30 | Hwang M. J., Jang A. S., Lim D. K., Sens. Actuator B: Chem., 2021, 329, 129134 |
| 31 | Lu S., Du J., Sun Z., Jing C., Anal. Chem., 2020, 92, 16229―16235 |
| 32 | Zhou H., Liu J., Xu J. J., Zhang S. S., Chen H. Y., Chem. Soc. Rev., 2018, 47, 1996―2019 |
| 33 | Feng Y., Zhou D., Gao L., He F., Biosens. Bioelectron., 2020, 168, 112527 |
| 34 | Lv M. M., Fan S. F., Wang Q. L., Lv Q. Y., Song X. J., Cui H. F., Microchim. Acta, 2020, 187, 73 |
| 35 | Liang T., Wu X., Chen B., Liu J., Aguilar Z. P., Xu H., LWT―Food Sci. Technol., 2020, 130, 109642 |
| 36 | Tang J., Wang Z. F., Zhou J. Q., Lu Q. J., Deng L., Spectroc. Acta Pt. A―Molec. Biomolec. Spectr., 2019, 215, 41―47 |
| 37 | Ali M., Li F., Zhang Z., Zhang K., Kang, D., Ankrum J., Le X., Zhao W., Chem. Soc. Rev., 2014, 43, 3324―3341 |
| 38 | Xu X., Su Y., Zhang Y., Wang X., Tian H., Ma X., Chu H., Xu W., Trac―Trends Anal. Chem., 2021, 141, 116293 |
| 39 | Li Y., Liu H., Huang H., Deng J., Fang L., Luo J., Zhang S., Huang J., Liang W., Zheng J., Biosens. Bioelectron., 2020, 147, 111752 |
| 40 | Yang K., Yu W. J., Huang G. R., Zhou J., Yang X., Fu W. L., RSC Adv., 2020, 10, 26824―26833 |
| 41 | Simmel F., Yurke B., Singh H., Chem. Rev., 2019, 119, 6326―6369 |
| 42 | Lu Y. Q., Luo F. F., Li Z., Dai G., Chu Z. H., Zhang J. W., Zhang F., Wang Q. J., He P. G., Talanta, 2021, 222, 121686 |
| 43 | Cai R. F., Yin, F., Zhang Z. W., Tian Y. P., Zhou N. D., Anal. Chim. Acta, 2019, 1075, 128―136 |
| 44 | Xiong E. H., Yan X. X., Zhang X. H., Li Y. M., Yang R. Y., Meng L. X., Chen J. H., Analyst, 2018, 143, 2799―2806 |
| 45 | Tao F., Fang J., Guo Y. C., Tao Y. Y., Han X. L., Hu Y. X., Wang J. J., Li L. Y., Jian Y. L., Xie G. M., Anal. Biochem., 2018, 554, 16―22 |
| 46 | Ning Z. Q., Zheng Y. J., Pan D., Zhang Y. J., Shen Y. F., Biosens. Bioelectron., 2020, 150, 7 |
| 47 | Leonardo S., ToldràA., Campàs M., Sensors, 2021, 21, 602 |
| 48 | Huang T. T., Liu S. C., Huang C. H., Lin C. J., Huang S. T., Electroanalysis, 2018, 30, 2397―2404 |
| 49 | Sharif S., Wang Y. X., Ye Z. Z., Wang Z., Qiu Q. M., Ying S. N., Ying Y. B., Sens. Actuator B: Chem., 2019, 301, 127051 |
| 50 | Nanayakkara I. A., White I. M., Analyst, 2019, 144, 3878―3885 |
| 51 | Takarada Y., Kodera T., Kobayashi K., Nakajima C., Kawase M., Suzuki Y., J. Microbiol. Methods, 2020, 177, 106062 |
| 52 | Liu M., Zhang Q., Chang D., Gu J., Brennan J. D., Li Y., Angew. Chem. Int. Ed., 2017, 56, 6142―6146 |
| 53 | Da-Silva E., Baudart J., Barthelmebs L., Talanta, 2018, 190, 410―422 |
| 54 | Chinnadayyala S. R., Park J., Le H. T. N., Santhosh M., Kadam A. N., Cho S., Biosens. Bioelectron., 2019, 126, 68―81 |
| 55 | Yang S., Yang C., Huang D., Song L., Chen J., Yang Q., Chem. Eur. J., 2019, 25, 5389―5405 |
| 56 | Ma X. Y., Ding W., Wang C., Wu H. J., Tian X. P., Lyu M. S., Wang S., J. Sens. Actuator B: Chem., 2021, 331, 129422 |
| 57 | Ali M. M., Wolfe M., Tram K., Gu J., Filipe C. D. M., Li Y. F., Brennan J. D., Angew. Chem. Int. Ed., 2019, 58, 9907―9911 |
| 58 | Ali M. M., Slepenkin A., Peterson E., Zhao W. A., ChemBioChem, 2019, 20, 906―910 |
| 59 | Shang Q. P., Su Y., Liang Y., Lai W., Jiang J., Wu H. Y., Zhang C. S., Anal. Bioanal. Chem., 2020, 412, 3787―3797 |
| 60 | Lee J. E., Mun H., Kim S. R., Kim M. G., Chang J. Y., Shim W. B., Biosens. Bioelectron., 2020, 151, 111968 |
| 61 | Zheng L. B., Qi P., Zhang D., Sens. Actuator B: Chem., 2018, 276, 42―47 |
| 62 | Mondal B., Bhavanashri N., Ramial S., Kingston J., J. Agric. Food Chem., 2018, 66, 1516―1522. |
| 63 | Wang D. X., Wang J., Du Y. C., Ma J. Y., Wang S. Y., Tang A. N., Kong D. M., Biosens. Bioelectron., 2020, 168, 112556 |
| 64 | Wu H., Chen X., Zhang M., Wang X., Chen Y., Qian C., Wu J., Xu J., Trac⁃Trends Anal. Chem., 2021, 135, 116150 |
| 65 | Bondy⁃Denomy J., Garcia B., Strum S., Du M. J., Rollins M. F., Hidalgo⁃Reyes Y., Wiedenheft B., Maxwell K. L., Davidson A. R., Nature, 2015, 526, 136―139 |
| 66 | Sheng A. Z., Wang, P., Yang J. Y., Tang L. F., Chen F., Zhang J., Anal. Chem., 2021, 93, 4676―4681 |
| 67 | Wang Y. F., Guo Y. C., Zhang L., Yang Y. J., Yang S. S., Yang L., Chen H. J., Liu C. G., Li J. J., Xie G. M., Sens. Actuator B: Chem., 2021, 334, 129600 |
| 68 | Peng L., Zhou J., Yin L. J., Man S. L., Ma L., Anal. Chim. Acta, 2020, 1125, 162―168 |
| 69 | Wang Y. Q., Ke Y. Q., Liu W. J., Sun Y. Q., Ding X. T., ACS Sens., 2020, 5, 1427―1435 |
| 70 | Zhou J., Yin L. J., Dong Y. N., Peng L., Liu G. Z., Man S. L., Ma L., Anal. Chim. Acta, 2020, 1127, 225―233 |
| 71 | Yan W. L., Gu L. D., Ren W., Ma X. Y., Qin M. C., Lyu M. S., Wang S. J., Helicobacter, 2019, 24, e12577 |
| 72 | Munzar J., Ng A., Juncker D., Chem. Soc. Rev., 2019, 48, 1390―1419 |
| 73 | Xie Y., Huang Y., Li J., Wu J., Sens. Actuator B: Chem., 2021, 339, 129865 |
| 74 | Trunzo N. E., Hong K. L., Int. J. Mol. Sci., 2020, 21, 5074 |
| 75 | Amraee M., Oloomi M., Yavari A., Bouzari S., Anal. Biochem., 2017, 536, 36―44 |
| 76 | Suh S. H., Choi S. J., Dwivedi H. P., Moore M. D., Escudero-Abarca B. I., Jaykus L. A., Anal. Biochem., 2018, 557, 27―33 |
| 77 | Ahn J. Y., Lee K. A., Lee M. J., Sekhon S. S., Rhee S. K., Cho S. J., Ko J. H., Lee L., Han J., Kim S. Y., Min J., Kim Y. H., J. Nanosci. Nanotechnol., 2018, 18, 1599―1605 |
| 78 | Cai R. F., Zhang Z. W., Chen H. H., Tian Y. P., Zhou N. D., Sens. Actuator B: Chem., 2021, 326, 128842 |
| 79 | Wu H. J., Gu L. D., Ma X. Y., Tian X. Q., Fan S. H., Qin M. C., Lu J., Lyu M. S., Wang S. J., ACS Omega, 2021, 6, 3771―3779 |
| 80 | Pla L., Santiago⁃Felipe S., Tormo⁃Mas M. A., Peman J., Sancenon F., Aznar E., Martinez⁃Manez R., Sens. Actuator B: Chem., 2020, 320, 128281 |
| 81 | Bayraç C., Eyidogan F., Oktem H. A., Biosens. Bioelectron., 2017, 98, 22―28 |
| 82 | Xu J. G., Guo J., Maina S. W., Yang Y. M., Hu Y. M., Li X. X., Qiu J. R., Xin Z. H., Anal. Biochem., 2018, 549, 136―142 |
| 83 | Li D., Yang E. L., Luo Z. W., Xie Q. Y., Duan Y. X., Nanoscale, 2021, 13, 2492―2501 |
| 84 | Jiang Y., Zou S., Cao X., Sens. Actuator B: Chem., 2017, 251, 976―984 |
| 85 | Li T., Ou G. Z., Chen X. L., Li Z. Y., Hu R., Li Y., Yang Y. H., Liu M. L., Anal. Chim. Acta, 2020, 1130, 20―28 |
| 86 | Jiang H., Sun Z., Guo Q., Weng X., Biosens Bioelectron., 2021, 182, 113191 |
| 87 | Liu Z. M., Yuan Y. Y., Wu X. Y., Ning Q. Q., Wu S. J., Fu L. Q., Sens. Actuator B: Chem., 2020, 322, 128646 |
| 88 | Zhang T., Zhou W. H., Lin X. Y., Khan M. R., Deng S., Zhou M., He G. P., Wu C. Y., Deng R. J., He Q., Biosens. Bioelectron., 2021, 176, 112906 |
| [1] | 王隆杰, 范鸿川, 秦渝, 曹秋娥, 郑立炎. 金属有机框架材料在分离分析领域的研究进展[J]. 高等学校化学学报, 2021, 42(4): 1167. |
| [2] | 刘珂, 靳宇, 梁建功, 吴园. 化学修饰提高核酸适体结合亲和力的研究进展[J]. 高等学校化学学报, 2021, 42(11): 3477. |
| [3] | 解忱, 陈娜, 杨雁冰, 袁荃. 核酸适体功能化的二维材料场效应晶体管传感器研究进展[J]. 高等学校化学学报, 2021, 42(11): 3406. |
| [4] | 赵卓, 王雪强. 核酸适体偶联药物的生物偶联构建技术与应用[J]. 高等学校化学学报, 2021, 42(11): 3367. |
| [5] | 刘学娇, 杨帆, 刘爽, 张春娟, 刘巧玲. 核酸适体靶向的膜蛋白识别与功能调控研究进展[J]. 高等学校化学学报, 2021, 42(11): 3277. |
| [6] | 刘园, 邓瑾琦, 赵帅, 田飞, 李轶, 孙佳姝, 刘超. 基于分子识别的免疫层析技术用于新冠肺炎感染的快速诊断[J]. 高等学校化学学报, 2021, 42(11): 3390. |
| [7] | 吉采灵, 程兴, 谈洁, 袁荃. 功能化核酸适体的筛选及分子识别应用[J]. 高等学校化学学报, 2021, 42(11): 3457. |
| [8] | 林宁钦, 姚克, 陈祥军. 晶状体蛋白识别互作与白内障的研究进展[J]. 高等学校化学学报, 2021, 42(11): 3379. |
| [9] | 黄玲, 庄梓健, 李翔, 石沐玲, 刘高强. 基于核酸适体的外泌体分子识别研究进展[J]. 高等学校化学学报, 2021, 42(11): 3493. |
| [10] | 李文帅, 武国瑞, 张茜菁, 岳爱琴, 杜维俊, 赵晋忠, 刘定斌. 基于拉曼光谱的细菌检测研究进展[J]. 高等学校化学学报, 2020, 41(5): 872. |
| [11] | 彭与煜,王煜,于鑫垚,曾巨澜,肖忠良,曹忠. 基于单(6-巯基-6-去氧)-β-环糊精修饰金电极对L-半胱氨酸的快速灵敏检测[J]. 高等学校化学学报, 2020, 41(2): 268. |
| [12] | 董倩, 李兆倩, 彭天欢, 陈卓, 谭蔚泓. 核酸适体在癌症诊疗中的研究进展[J]. 高等学校化学学报, 2020, 41(12): 2648. |
| [13] | 马玉聪, 樊保民, 郝华, 吕金玉, 冯云皓, 杨彪. 十八胺基分子组装体在碳钢表面的作用机理与模拟[J]. 高等学校化学学报, 2019, 40(1): 96. |
| [14] | 沈晓琴, 李智, 王刚林, 王莉, 孙权洪, 罗序成, 马楠. 基于氧化石墨烯和DNA量子点组装体的DNA二元逻辑检测及循环可逆设计[J]. 高等学校化学学报, 2017, 38(12): 2176. |
| [15] | 宋春霞, 羊小海, 王柯敏, 王青, 刘剑波, 黄晋, 李文山, 黄海花, 刘卫. 聚合物在荧光检测领域的应用[J]. 高等学校化学学报, 2016, 37(2): 201. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||