

高等学校化学学报 ›› 2021, Vol. 42 ›› Issue (11): 3457.doi: 10.7503/cjcu20210442
收稿日期:2021-06-28
出版日期:2021-11-10
发布日期:2021-11-10
通讯作者:
袁荃
E-mail:tanjie0416@hnu.edu.cn;yuanquan@whu.edu.cn
作者简介:谈 洁, 女, 博士, 副教授, 主要从事化学修饰核酸的设计及其生物医学应用研究. E-mail: 基金资助:
JI Cailing, CHENG Xing, TAN Jie( ), YUAN Quan(
), YUAN Quan( )
)
Received:2021-06-28
Online:2021-11-10
Published:2021-11-10
Contact:
YUAN Quan
E-mail:tanjie0416@hnu.edu.cn;yuanquan@whu.edu.cn
Supported by:摘要:
核酸适体是从寡核苷酸文库中筛选获得的一段单链寡核苷酸. 由于能与多种靶标分子高特异性结合, 核酸适体已发展成为一种新兴的分子识别工具, 广泛应用于生物医学等领域. 天然核酸文库有限的化学组成限制了核酸适体的结构和功能, 进而限制了其在分子识别中的应用. 功能化核酸适体通过引入特定的化学官能团使核酸序列具有更丰富的构象和功能, 增强其分子识别能力. 然而, 功能化核酸很难与核酸扩增方法兼容, 因而难以使用传统筛选方法进行功能化核酸的筛选. 因此, 优化筛选方法对于获得具有优异性能的功能化核酸适体至关重要. 本综述总结了功能化核酸适体的筛选方法, 并介绍了其作为分子识别工具在生物医学领域中的应用.
中图分类号:
TrendMD:
吉采灵, 程兴, 谈洁, 袁荃. 功能化核酸适体的筛选及分子识别应用. 高等学校化学学报, 2021, 42(11): 3457.
JI Cailing, CHENG Xing, TAN Jie, YUAN Quan. Selection of Functionalized Aptamers and Their Applications in Molecular Recognition. Chem. J. Chinese Universities, 2021, 42(11): 3457.
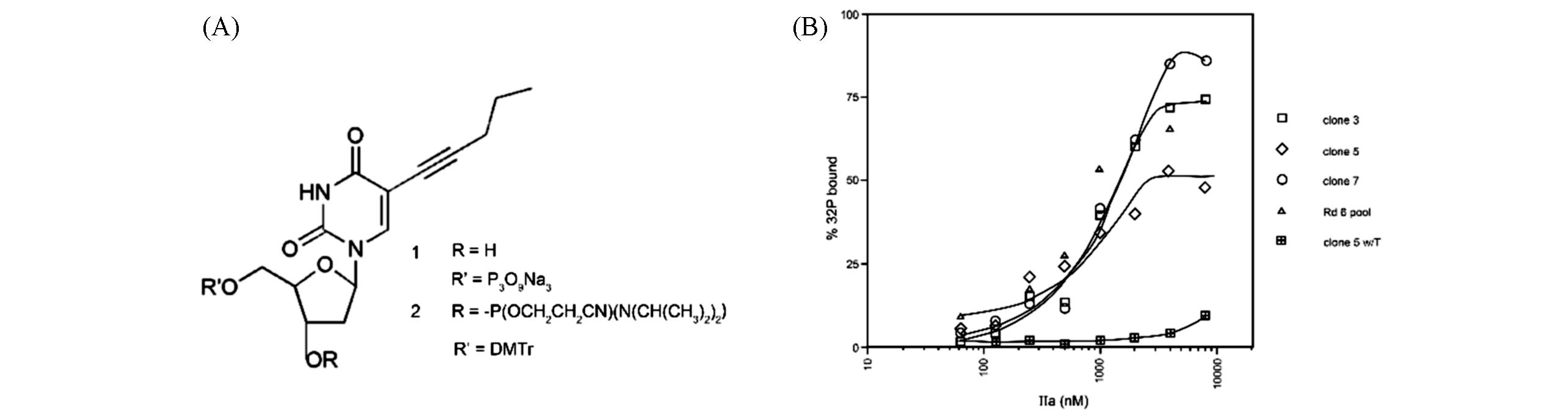
Fig.1 Chemical structures of 5?(1?pentynyl)?2′?deoxyuridine(A) and the interaction between radiolabelled aptamers and thrombin evaluated by itrocellulose filter binding(B)[28]Copyright 1994, Oxford University Press.
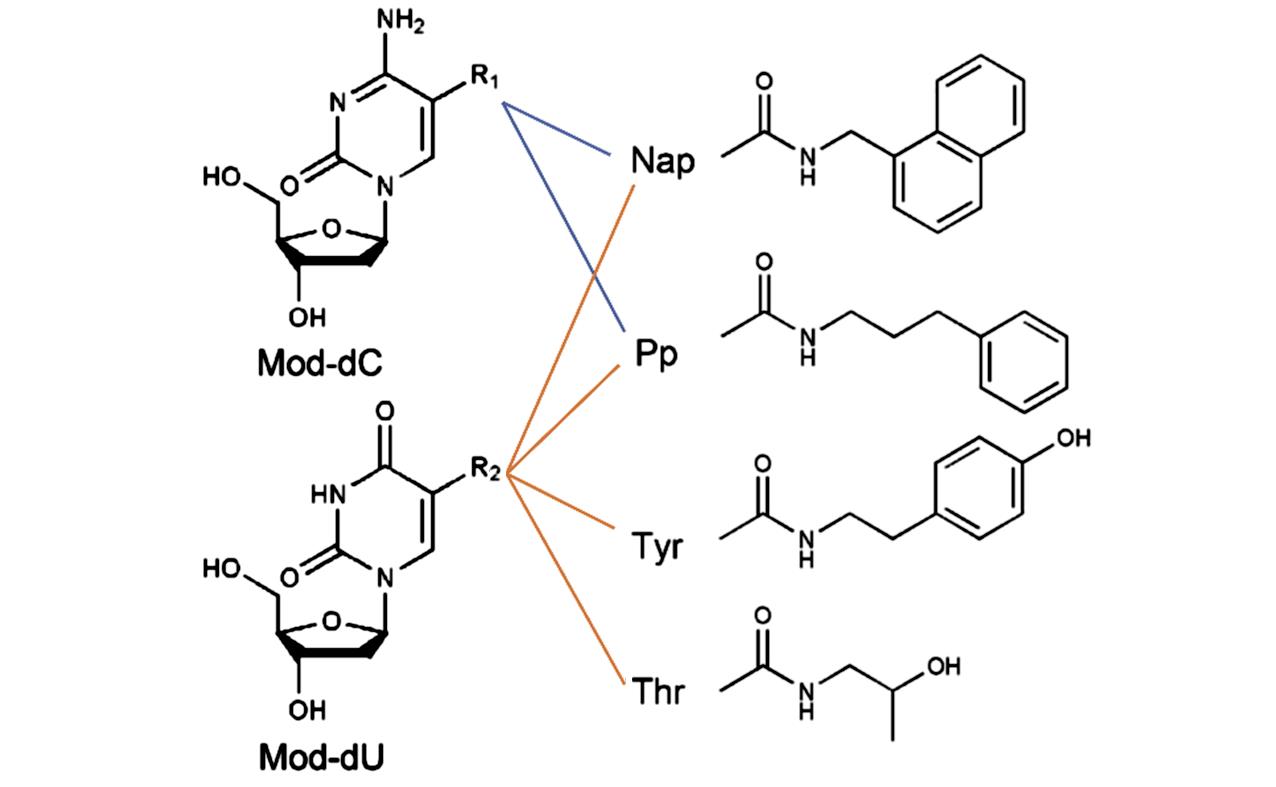
Fig.2 Chemical structures of modified deoxycytidine(Mod?dC) and modified deoxyuridine(Mod?dU) bearing a 5?(N?substituted?carboxamide) functional group R1 and R2, respectivelyR1 group: Modifications on dC; R2 group: modifications on dU.
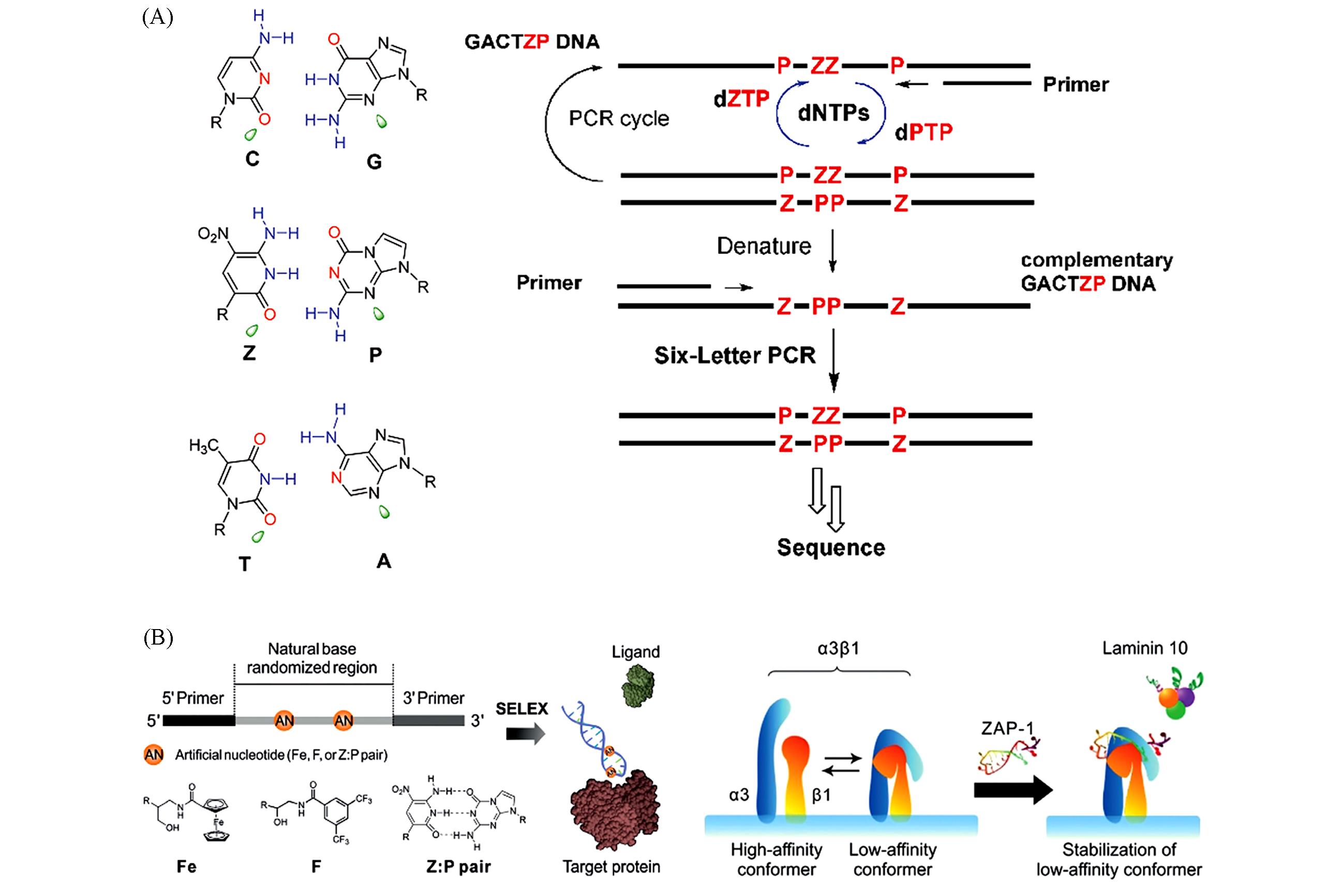
Fig.3 GACTZP artificial genetic system(A)[33], schematic diagram of molecular design of aptamers containing P?Z pairs and the conformational regulation of integrin α3β1(B)[15](A) Left column: matched C?G, P?Z, and T?A pairs. Green parts: electron density presented to the minor groove. Right column: the screening process by the six?letter artificial genetic system. (A) Copyright 2011, American Chemical Society; (B) Copyright 2019, Wiley?VCH.
| Type of accepted modification | Functional aptamers were screened using modified nucleic acid library | Ref. |
|---|---|---|
| 2′?OMe | Human neutrophil elastase(HNE) aptamer | [ |
| 2′?Fluoro | Human neutrophil elastase(HNE) aptamer | [ |
| 2′?Azido | Blood coagulation protein factor IX aptamer, neutrophil elastase(HNE) aptamer | [ |
| 2′?Chloro, 2′?amino/arabinose sugars | —— | [ |
Table 1 Modified substrates of SFM4-3 polymerase
| Type of accepted modification | Functional aptamers were screened using modified nucleic acid library | Ref. |
|---|---|---|
| 2′?OMe | Human neutrophil elastase(HNE) aptamer | [ |
| 2′?Fluoro | Human neutrophil elastase(HNE) aptamer | [ |
| 2′?Azido | Blood coagulation protein factor IX aptamer, neutrophil elastase(HNE) aptamer | [ |
| 2′?Chloro, 2′?amino/arabinose sugars | —— | [ |
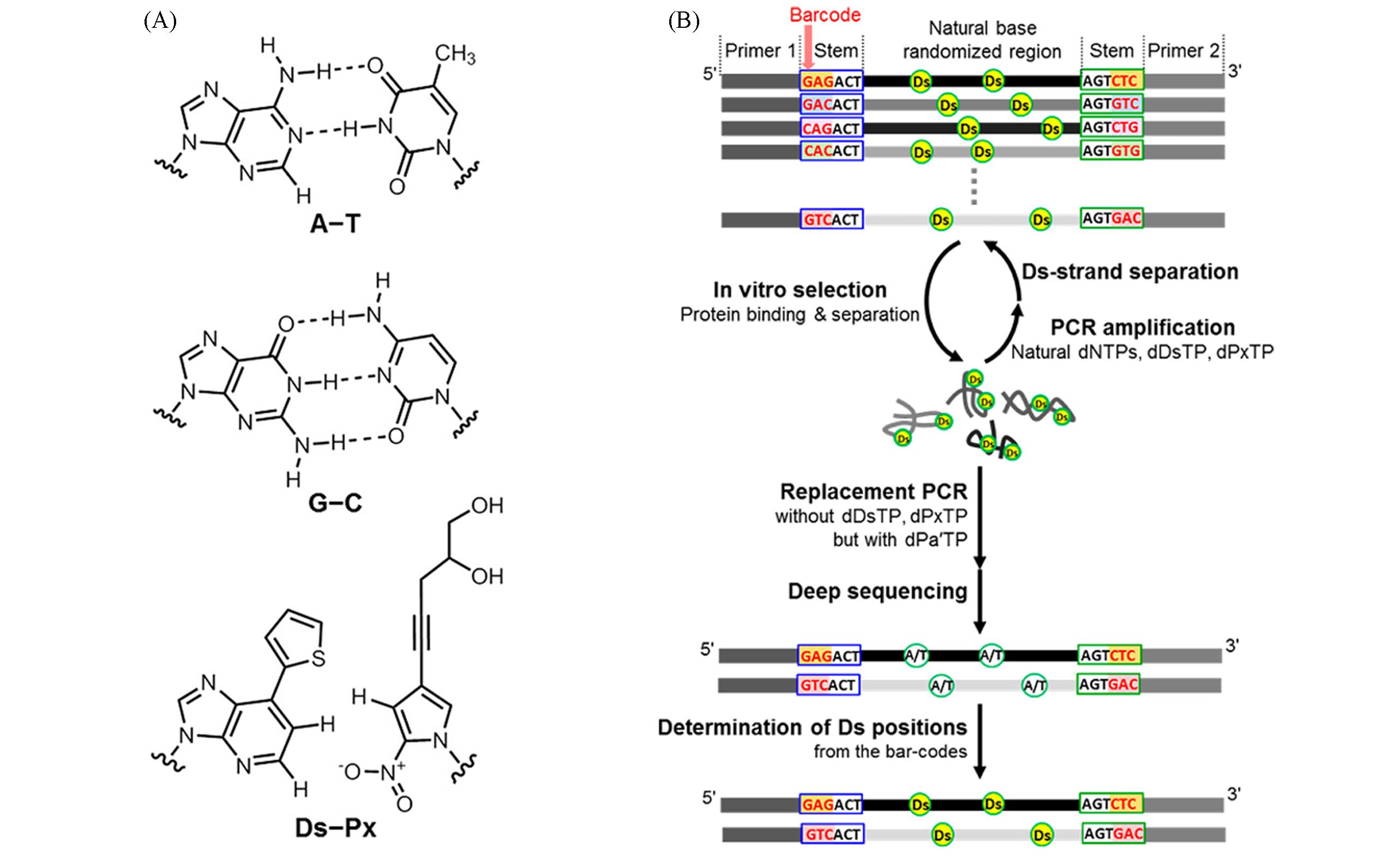
Fig.4 Chemical structures of base pairs in DNA libraries containing hydrophobic Ds bases(A)[45] and scheme of the SELEX procedure using DNA libraries with five different bases(B)[46](A) Copyright 2019, American Chemical Society; (B) Copyright 2017, American Chemical Society.
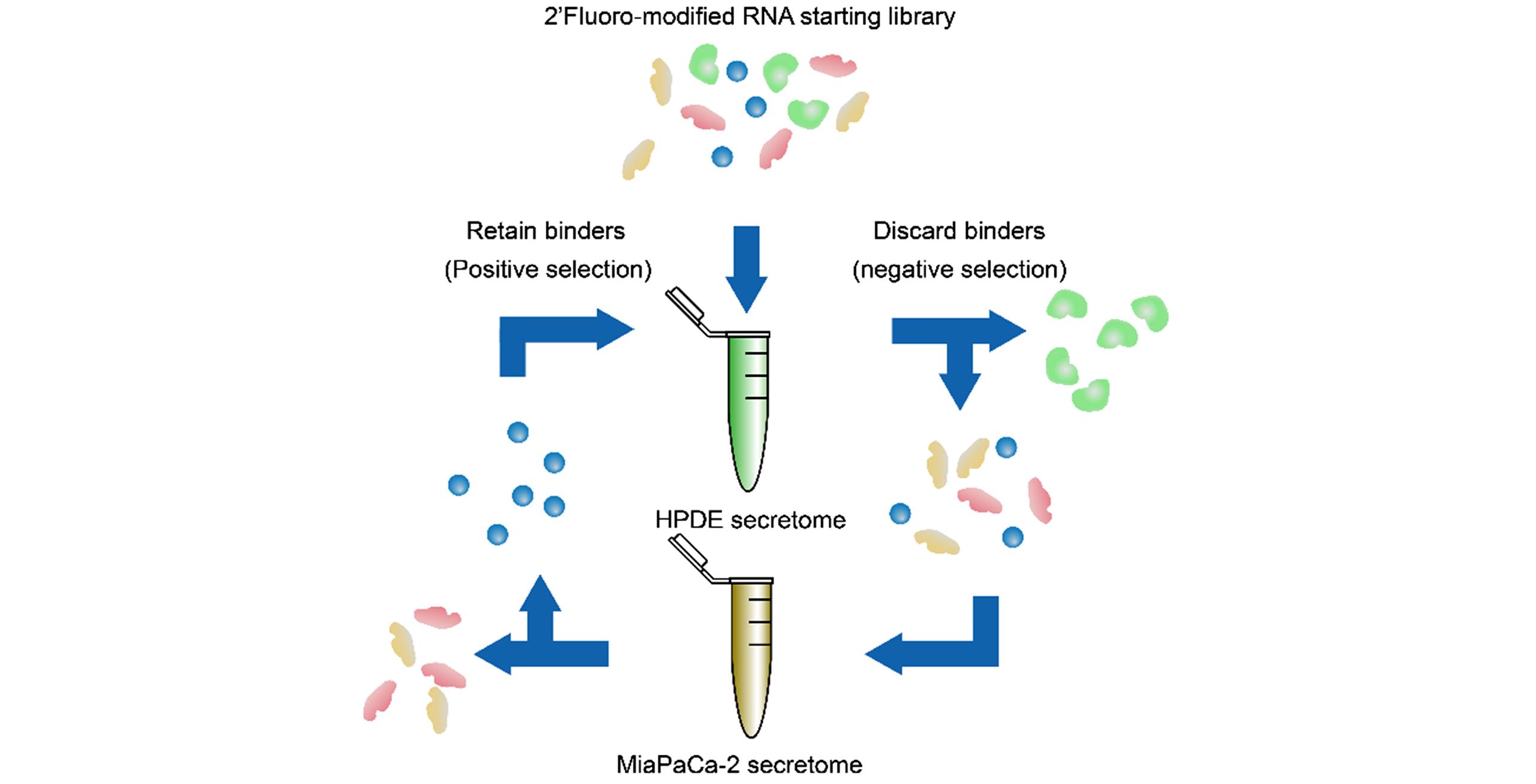
Fig.5 Schemeo of “positive/negative” SELEX strategyPositive selection: human pancreatic ductal adenocarcinoma cell line MiaPaCa?2; negative selection: human pancreatic ductal epithelial cell line HPDE?E6E7.
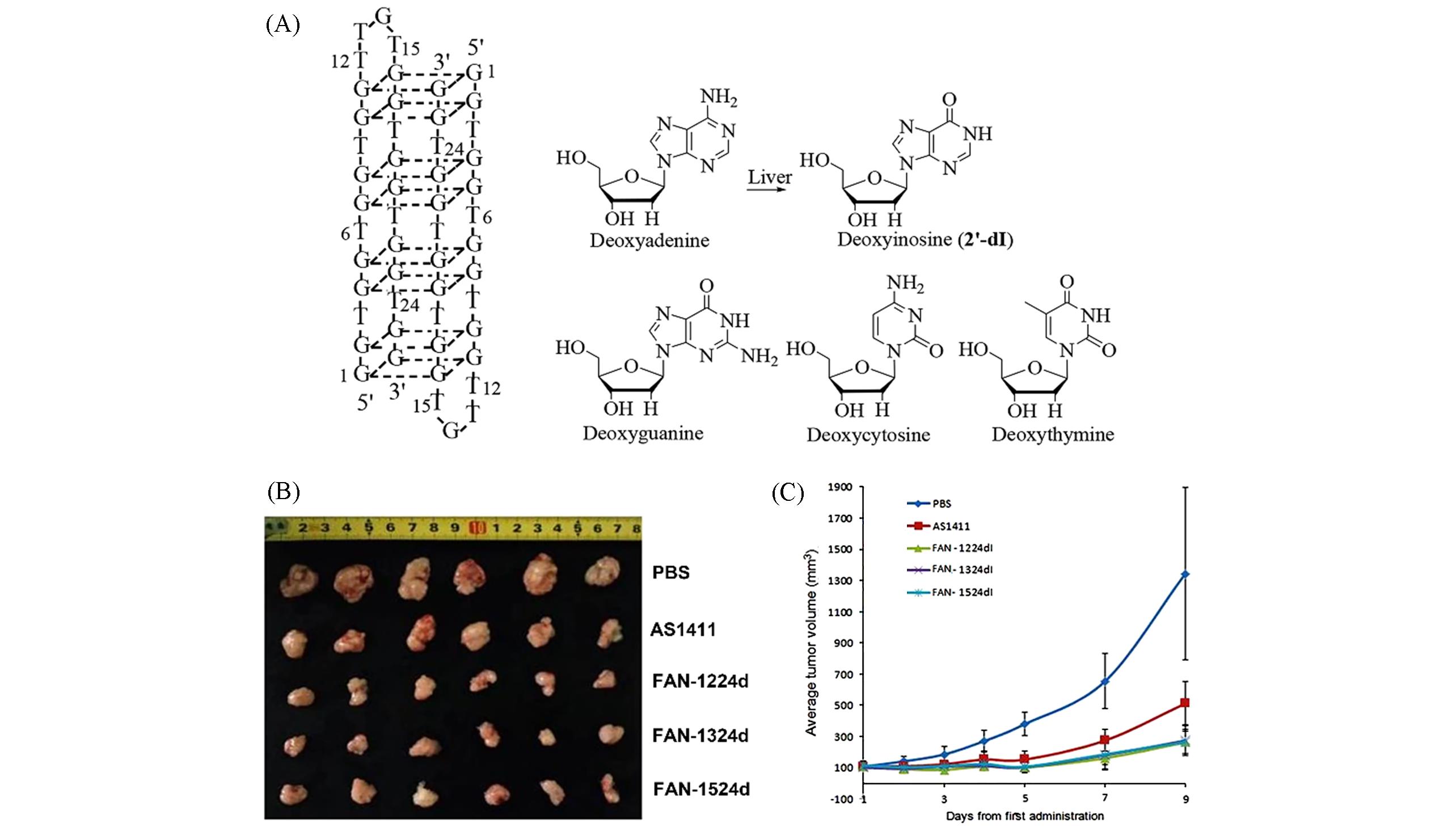
Fig.6 The structure diagram of 2′?deoxynucleosides and AS1411(A), photographs of the dissected tumors after treated with AS1411 or functionalized AS1411(B) and average tumor volume after treated with AS1411 or functionalized AS1411(C)[77]Copyright 2016, Springer Nature.
| Aptamer | Sequence(5′?3′) | Aptamer | Sequence(5′?3′) |
|---|---|---|---|
| AS1411 | ggt ggt ggt ggt tgt ggt ggt ggt gg | FAN?1324dI | ggt ggt ggt ggt dIgt ggt ggt ggdI gg |
| FAN?1224dI | ggt ggt ggt ggdI tgt ggt ggt ggdI gg | FAN?1524dI | ggt ggt ggt ggt tgdI ggt ggt ggdI gg |
Table 2 Sequences of AS1411 and 2′-dI modified AS1411s
| Aptamer | Sequence(5′?3′) | Aptamer | Sequence(5′?3′) |
|---|---|---|---|
| AS1411 | ggt ggt ggt ggt tgt ggt ggt ggt gg | FAN?1324dI | ggt ggt ggt ggt dIgt ggt ggt ggdI gg |
| FAN?1224dI | ggt ggt ggt ggdI tgt ggt ggt ggdI gg | FAN?1524dI | ggt ggt ggt ggt tgdI ggt ggt ggdI gg |
| Aptamer | KD/(nmol·L-1) | Aptamer | KD/(nmol·L-1) |
|---|---|---|---|
| AS1411 | 148.0 | FCL?I(6L /12D) | 9.56 |
| 12D | 31.9 | FCL?II(6L /12D/24dI) | 4.7 |
Table 3 Binding parameters for the affinity of D-/L-isoT(TD/TL) and/or 2′-dI modified AS1411 for nucleolin
| Aptamer | KD/(nmol·L-1) | Aptamer | KD/(nmol·L-1) |
|---|---|---|---|
| AS1411 | 148.0 | FCL?I(6L /12D) | 9.56 |
| 12D | 31.9 | FCL?II(6L /12D/24dI) | 4.7 |
| 1 | Ng E. W., Shima D. T., Calias P., Cunningham E. T. Jr., Guyer D. R., Adamis A. P., Nat. Rev. Drug Discov.,2006, 5(2), 123—132 |
| 2 | Sefah K., Shangguan D., Xiong X., O’Donoghue M. B., Tan W., Nat. Protoc.,2010, 5(6), 1169—1185 |
| 3 | Mayer G., Angew. Chem. Int. Ed.,2009, 48(15), 2672—2689 |
| 4 | Russo Krauss I., Spiridonova V., Pica A., Napolitano V., Sica F., Nucleic Acids Res.,2016, 44(2), 983—991 |
| 5 | Hermann T., Patel D. J., Science,2000, 287(5454), 820—825 |
| 6 | Mascini M., Palchetti I., Tombelli S., Angew. Chem. Int. Ed.,2012, 51(6), 1316—1332 |
| 7 | Opazo F., Levy M., Byrom M., Schafer C., Geisler C., Groemer T. W., Ellington A. D., Rizzoli S. O., Nat. Methods,2012, 9(10), 938—939 |
| 8 | Zhu G., Ye M., Donovan M. J., Song E., Zhao Z., Tan W., Chem. Commun.,2012, 48(85), 10472—10480 |
| 9 | Guan B., Zhang X., Int. J. Nanomed.,2020, 15, 1059—1071 |
| 10 | Zhou J., Rossi J. J., Mol. Ther.—Nucleic Acids,2014, 3, e169 |
| 11 | Li L., Xu S., Yan H., Li X., Yazd H. S., Li X., Huang T., Cui C., Jiang J., Tan W., Angew. Chem. Int. Ed.,2021, 60(5), 2221—2231 |
| 12 | Sun H., Zhu X., Lu P. Y., Rosato R. R., Tan W., Zu Y., Mol. Ther.—Nucleic Acids,2014, 3, e182 |
| 13 | Sun H., Zu Y., Small,2015, 11(20), 2352—2364 |
| 14 | Gawande B. N., Rohloff J. C., Carter J. D., von Carlowitz I., Zhang C., Schneider D. J., Janjic N., Proc. Natl. Acad. Sci. USA,2017, 114(11), 2898—2903 |
| 15 | Tan J., Zhao M., Wang J., Li Z., Liang L., Zhang L., Yuan Q., Tan W., Angew. Chem. Int. Ed.,2019, 58(6), 1621—1625 |
| 16 | Li L., Xu S., Yan H., Li X., Yazd H. S., Li X., Huang T., Cui C., Jiang J., Tan W., Angew. Chem. Int. Ed. Engl.,2021, 60(5), 2221—2231 |
| 17 | Lipi F., Chen S., Chakravarthy M., Rakesh S., Veedu R. N., RNA Biol.,2016, 13(12), 1232—1245 |
| 18 | Shen R., Tan J., Yuan Q., ACS Appl. Bio Mater.,2020, 3(5), 2816—2826 |
| 19 | Tolle F., Brandle G. M., Matzner D., Mayer G., Angew. Chem. Int. Ed.,2015, 54(37), 10971—10974 |
| 20 | Kimoto M., Yamashige R., Matsunaga K., Yokoyama S., Hirao I., Nat. Biotechnol.,2013, 31(5), 453—457 |
| 21 | Eissa S., Siddiqua A., Chinnappan R., Zourob M., ACS Appl. Bio Mater.,2019, 2(6), 2624—2632 |
| 22 | Zhuo Z., Yu Y., Wang M., Li J., Zhang Z., Liu J., Wu X., Lu A., Zhang G., Zhang B., Int. J. Mol. Sci.,2017, 18(10), 2142 |
| 23 | Zhu Z., Song Y., Li C., Zou Y., Zhu L., An Y., Yang C. J., Anal. Chem.,2014, 86(12), 5881—5888 |
| 24 | Da Rocha Gomes S., Miguel J., Azema L., Eimer S., Ries C., Dausse E., Loiseau H., Allard M., Toulme J. J., Bioconjug. Chem.,2012, 23(11), 2192—2200 |
| 25 | Gordon C. K. L., Wu D., Pusuluri A., Feagin T. A., Csordas A. T., Eisenstein M. S., Hawker C. J., Niu J., Soh H. T., ACS Chem. Biol.,2019, 14(12), 2652—2662 |
| 26 | Duo J., Chiriac C., Huang R. Y., Mehl J., Chen G., Tymiak A., Sabbatini P., Pillutla R., Zhang Y., Anal. Chem.,2018, 90(8), 5162—5170 |
| 27 | Tolle F., Rosenthal M., Pfeiffer F., Mayer G., Bioconjug. Chem.,2016, 27(3), 500—503 |
| 28 | Latham J. A., Johnson R., Toole J. J., Nucleic Acids Res.,1994, 22(14), 2817—2822 |
| 29 | Lauridsen L. H., Rothnagel J. A., Veedu R. N., ChemBioChem,2012, 13(1), 19—25 |
| 30 | Vaught J. D., Bock C., Carter J., Fitzwater T., Otis M., Schneider D., Rolando J., Waugh S., Wilcox S. K., Eaton B. E., J. Am. Chem. Soc.,2010, 132(12), 4141—4151 |
| 31 | Arca M., Recenti Prog. Med.,2019, 110(9), 401—411 |
| 32 | Cariou B., Le May C., Costet P., Atherosclerosis,2011, 216(2), 258—265 |
| 33 | Yang Z., Chen F., Alvarado J. B., Benner S. A., J. Am. Chem. Soc.,2011, 133(38), 15105—15112 |
| 34 | Pfeiffer F., Rosenthal M., Siegl J., Ewers J., Mayer G., Curr. Opin. Biotechnol.,2017, 48, 111—118 |
| 35 | Joyce C. M., Steitz T. A., Annu. Rev. Biochem.,1994, 63, 777—822 |
| 36 | Chen T., Hongdilokkul N., Liu Z., Adhikary R., Tsuen S. S., Romesberg F. E., Nat. Chem.,2016, 8(6), 556—562 |
| 37 | Chen T., Romesberg F. E., J. Am. Chem. Soc.,2017, 139(29), 9949—9954 |
| 38 | Chen T., Romesberg F. E., Angew. Chem. Int. Ed.,2017, 56(45), 14046—14051 |
| 39 | Liu Z., Chen T., Romesberg F. E., Chem. Sci.,2017, 8(12), 8179—8182 |
| 40 | Shao Q., Chen T., Sheng K., Liu Z., Zhang Z., Romesberg F. E., J. Am. Chem. Soc.,2020, 142(5), 2125—2128 |
| 41 | Thirunavukarasu D., Chen T., Liu Z., Hongdilokkul N., Romesberg F. E., J. Am. Chem. Soc.,2017, 139(8), 2892—2895 |
| 42 | Hirao I., Kimoto M., Lee K. H., Biochimie,2018, 145, 15—21 |
| 43 | Dien V. T., Morris S. E., Karadeema R. J., Romesberg F. E., Curr. Opin. Chem. Biol.,2018, 46, 196—202 |
| 44 | Kimoto M., Kawai R., Mitsui T., Yokoyama S., Hirao I., Nucleic Acids Res.,2009, 37(2), e14 |
| 45 | Hamashima K., Soong Y. T., Matsunaga K. I., Kimoto M., Hirao I., ACS Synth. Biol.,2019, 8(6), 1401—1410 |
| 46 | Matsunaga K. I., Kimoto M., Hirao I., J. Am. Chem. Soc.,2017, 139(1), 324—334 |
| 47 | Zhang Z., Liu J., ACS Appl. Mater. Interfaces,2016, 8(10), 6371—6378 |
| 48 | Fukunishi H., Shimada J., Shiraishi K., Biochem.,2012, 51(12), 2597—2605 |
| 49 | Qu L., Qiao X., Qi F., Nishida N., Hoshino T., J. Chem. Inf. Model.,2021, 61(5), 2396—2406 |
| 50 | Zhou J., Battig M. R., Wang Y., Anal. Bioanal. Chem.,2010, 398(6), 2471—2480 |
| 51 | Pashchenko O., Shelby T., Banerjee T., Santra S., ACS Infect. Dis.,2018, 4(8), 1162—1178 |
| 52 | Kang S., Hah S. S., Bioconjug. Chem.,2014, 25(8), 1421—1427 |
| 53 | Drabik A., Ner⁃Kluza J., Mielczarek P., Civit L., Mayer G., Silberring J., J. Proteome Res.,2018, 17(6), 2174—2181 |
| 54 | Ni S., Zhuo Z., Pan Y., Yu Y., Li F., Liu J., Wang L., Wu X., Li D., Wan Y., Zhang L., Yang Z., Zhang B. T., Lu A., Zhang G., ACS Appl. Mater. Interfaces,2021, 13(8), 9500—9519 |
| 55 | Wang J., Fang X., Zhang C., Ji H., Pang Q., Li X., Luo Z., Wu Q., Zhang L., ACS Appl. Mater. Interfaces,2021, 13(14), 16118—16126 |
| 56 | Yi K., Rong Y., Huang L., Tang X., Zhang Q., Wang W., Wu J., Wang F., ACS Sens.,2021, 6(4), 1418—1429 |
| 57 | Canoura J., Yu H., Alkhamis O., Roncancio D., Farhana R., Xiao Y., J. Am. Chem. Soc.,2021, 143(2), 805—816 |
| 58 | Huang J., Chen X. X., Fu X. K., Li Z., Huang Y. H., Liang C., Front. Cell Dev. Biol.,2021, 9, 659760 |
| 59 | Califf R. M., Exp. Biol. Med.,2018, 243(3), 213—221 |
| 60 | Shangguan D., Cao Z. H., Meng L., Mallikaratchy P., Sefah K., Wang H., Li Y., Tan W. H., J. Proteome Res.,2008, 7(5), 2133—2139 |
| 61 | Werner J., Combs S. E., Springfeld C., Hartwig W., Hackert T., Buchler M. W., Nat. Rev. Clin. Oncol.,2013, 10(6), 323—333 |
| 62 | Ray P., Rialon⁃Guevara K. L., Veras E., Sullenger B. A., White R. R., J. Clin. Invest.,2012, 122(5), 1734—1741 |
| 63 | Liu B., Bell A. W., Paranjpe S., Bowen W. C., Khillan J. S., Luo J. H., Mars W. M., Michalopoulos G. K., Hepatology,2010, 52(3), 1060—1067 |
| 64 | Liu X., Mao D., Deng G., Song Y., Zhang F., Yang S., Li G., Liu F., Cao W., Zhu X., Theranostics,2020, 10(10), 4410—4421 |
| 65 | Imai K., Hirata S., Irie A., Senju S., Ikuta Y., Yokomine K., Harao M., Inoue M., Tsunoda T., Nakatsuru S., Nakagawa H., Nakamura Y., Baba H., Nishimura Y., Clin. Cancer. Res.,2008, 14(20), 6487—6495 |
| 66 | Kumar S., Mohan A., Guleria R., Biomarkers,2006, 11(5), 385—405 |
| 67 | Shukla H. D., Vaitiekunas P., Cotter R. J., Proteomics,2012, 12(19/20), 3085—104 |
| 68 | Zhong L., Liu Y., Wang K., He Z., Gong Z., Zhao Z., Yang Y., Gao X., Li F., Wu H., Zhang S., Chen L., BMC Cancer,2018, 18(1), 911 |
| 69 | Mallikaratchy P., Tang Z., Kwame S., Meng L., Shangguan D., Tan W., Mol. Cell. Proteomics,2007, 6(12), 2230—2238 |
| 70 | Kerbel R. S., Bioessays,1991, 13(1), 31—36 |
| 71 | Creixell M., Peppas N. A., Nano Today,2012, 7(4), 367—379 |
| 72 | Wang J., Wei Y., Hu X., Fang Y. Y., Li X., Liu J., Wang S., Yuan Q., J. Am. Chem. Soc.,2015, 137(33), 10576—10584 |
| 73 | Thomas C. J., Rahier N. J., Hecht S. M., Bioorg. Med. Chem.,2004, 12(7), 1585—1604 |
| 74 | Imaizumi Y., Kasahara Y., Fujita H., Kitadume S., Ozaki H., Endoh T., Kuwahara M., Sugimoto N., J. Am. Chem. Soc.,2013, 135(25), 9412—9419 |
| 75 | Trujillo C. A., Nery A. A., Alves J. M., Martins A. H., Ulrich H., Clin. Ophthalmol.,2007, 1(4), 393—402 |
| 76 | Gutsaeva D. R., Parkerson J. B., Yerigenahally S. D., Kurz J. C., Schaub R. G., Ikuta T., Head C. A., Blood,2011, 117(2), 727—735 |
| 77 | Fan X., Sun L., Wu Y., Zhang L., Yang Z., Sci. Rep.,2016, 6(1), 25799 |
| 78 | Fan X., Sun L., Li K., Yang X., Cai B., Zhang Y., Zhu Y., Ma Y., Guan Z., Wu Y., Zhang L., Yang Z., Mol. Ther.—Nucleic Acids,2017, 9, 218—229 |
| 79 | Ruddle N. H., J. Immunol.,1986, 136(6), 2335—2336 |
| 80 | Shalaby M. R., Aggarwal B. B., Rinderknecht E., Svedersky L. P., Finkle B. S., Palladino M. A. Jr., J. Immunol.,1985, 135(3), 2069—2073 |
| 81 | Matsunaga K., Kimoto M., Hanson C., Sanford M., Young H. A., Hirao I., Sci. Rep.,2015, 5, 18478 |
| [1] | 王隆杰, 范鸿川, 秦渝, 曹秋娥, 郑立炎. 金属有机框架材料在分离分析领域的研究进展[J]. 高等学校化学学报, 2021, 42(4): 1167. |
| [2] | 林宁钦, 姚克, 陈祥军. 晶状体蛋白识别互作与白内障的研究进展[J]. 高等学校化学学报, 2021, 42(11): 3379. |
| [3] | 黄玲, 庄梓健, 李翔, 石沐玲, 刘高强. 基于核酸适体的外泌体分子识别研究进展[J]. 高等学校化学学报, 2021, 42(11): 3493. |
| [4] | 张晓荣, 陈岚岚, 胡善文. 基于分子识别的细菌检测研究进展[J]. 高等学校化学学报, 2021, 42(11): 3468. |
| [5] | 刘学娇, 杨帆, 刘爽, 张春娟, 刘巧玲. 核酸适体靶向的膜蛋白识别与功能调控研究进展[J]. 高等学校化学学报, 2021, 42(11): 3277. |
| [6] | 刘园, 邓瑾琦, 赵帅, 田飞, 李轶, 孙佳姝, 刘超. 基于分子识别的免疫层析技术用于新冠肺炎感染的快速诊断[J]. 高等学校化学学报, 2021, 42(11): 3390. |
| [7] | 叶成濠, 梁亨, 李恩民, 许丽艳, 李鹏, 陈广慧. 高通量虚拟筛选CDK2/Cyclin A2靶点抑制剂[J]. 高等学校化学学报, 2021, 42(10): 3135. |
| [8] | 彭与煜,王煜,于鑫垚,曾巨澜,肖忠良,曹忠. 基于单(6-巯基-6-去氧)-β-环糊精修饰金电极对L-半胱氨酸的快速灵敏检测[J]. 高等学校化学学报, 2020, 41(2): 268. |
| [9] | 李显明, 郑婷, 高露, 李峰, 侯贤灯, 吴鹏. 重组酶聚合酶扩增:扩增原理及性能分析[J]. 高等学校化学学报, 2020, 41(12): 2587. |
| [10] | 佟宗轩, 胡沁沁, 顾宏周. DNA酶: 筛选、 生物传感及展望[J]. 高等学校化学学报, 2020, 41(11): 2345. |
| [11] | 张志庆,汪珊珊,张子辰,马杰,王秀凤,周亭,王芳,张国栋. 基于滚环合成的聚分子信标用于凝血酶的靶向检测[J]. 高等学校化学学报, 2019, 40(12): 2465. |
| [12] | 王洁,张宇,于敏,方瑾. 基于双向浓度梯度的微流控芯片系统对肿瘤细胞侵袭能力的多重分析[J]. 高等学校化学学报, 2019, 40(12): 2494. |
| [13] | 刘中成, 刘世芳, 张苏, 杨艳蕾, 李飞, 张楠, 袁欣, 张艳芬. Cε3-Cε4蛋白寡核苷酸适配子结合位点的预测与筛选[J]. 高等学校化学学报, 2019, 40(1): 83. |
| [14] | 马玉聪, 樊保民, 郝华, 吕金玉, 冯云皓, 杨彪. 十八胺基分子组装体在碳钢表面的作用机理与模拟[J]. 高等学校化学学报, 2019, 40(1): 96. |
| [15] | 刘宇博, 张娜娜, 陈锦娇, 朱彤, 张嘉宁, 李文利. O-连接N-乙酰葡糖胺转移酶抑制剂的筛选与活性分析[J]. 高等学校化学学报, 2018, 39(6): 1185. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||