

高等学校化学学报 ›› 2020, Vol. 41 ›› Issue (12): 2648.doi: 10.7503/cjcu20200436
• 庆祝《高等学校化学学报》复刊40周年专栏 • 上一篇 下一篇
董倩1,2, 李兆倩1,2, 彭天欢1,2, 陈卓1,2( ), 谭蔚泓1,2,3(
), 谭蔚泓1,2,3( )
)
收稿日期:2020-07-08
出版日期:2020-12-10
发布日期:2020-12-09
通讯作者:
陈卓
E-mail:zhuochen@hnu.edu.cn;tan@hnu.edu.cn
作者简介:谭蔚泓, 男, 博士, 教授, 中国科学院院士, 主要从事生物分析化学、 化学生物学和分子医学方面的研究. E-mail: 基金资助:
DONG Qian1,2, LI Zhaoqian1,2, PENG Tianhuan1,2, CHEN Zhuo1,2( ), TAN Weihong1,2,3(
), TAN Weihong1,2,3( )
)
Received:2020-07-08
Online:2020-12-10
Published:2020-12-09
Contact:
CHEN Zhuo
E-mail:zhuochen@hnu.edu.cn;tan@hnu.edu.cn
Supported by:摘要:
核酸适体是通过体外筛选技术得到的可特异性结合靶标分子的单链寡核苷酸分子探针, 其表现出与抗体相当或更优异的特异性和亲和力, 且具有靶标范围广、 免疫原性低、 易于精准制备和修饰及设计灵活可控等优势. 为癌症的早期筛查、 诊断及靶向治疗提供了全新的分子工具, 在癌症诊疗领域获得了广泛的关注与应用. 本文聚焦核酸适体在癌症诊断及治疗中的应用, 对近年来取得的研究进展进行了系统性总结, 并对未来发展方向及前景进行了展望.
中图分类号:
TrendMD:
董倩, 李兆倩, 彭天欢, 陈卓, 谭蔚泓. 核酸适体在癌症诊疗中的研究进展. 高等学校化学学报, 2020, 41(12): 2648.
DONG Qian, LI Zhaoqian, PENG Tianhuan, CHEN Zhuo, TAN Weihong. Progress on Aptamer for Cancer Theranostics. Chem. J. Chinese Universities, 2020, 41(12): 2648.
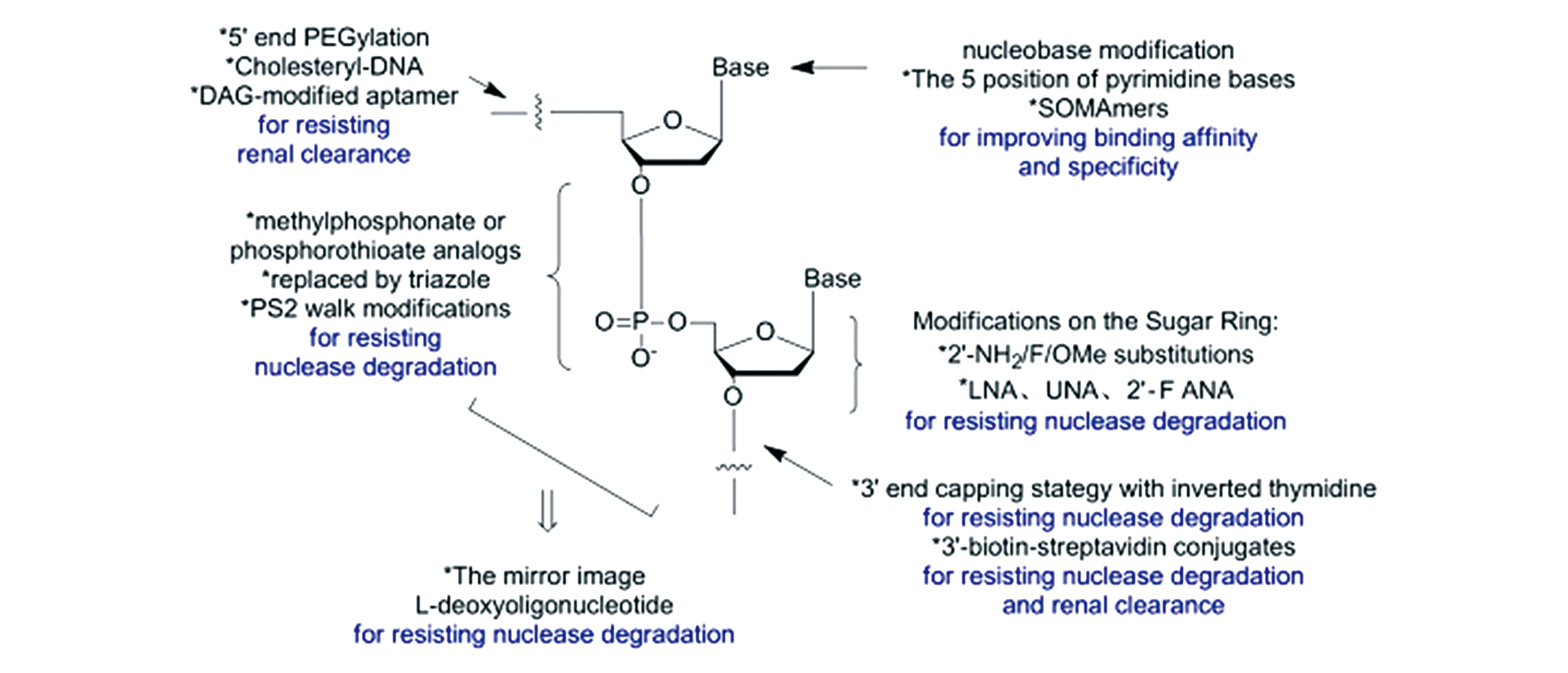
Fig.2 Common strategies in the chemical modifications of nucleic acid aptamers and their purposes[64]Copyright 2007, Multidisciplinary Digital Publishing Institute.
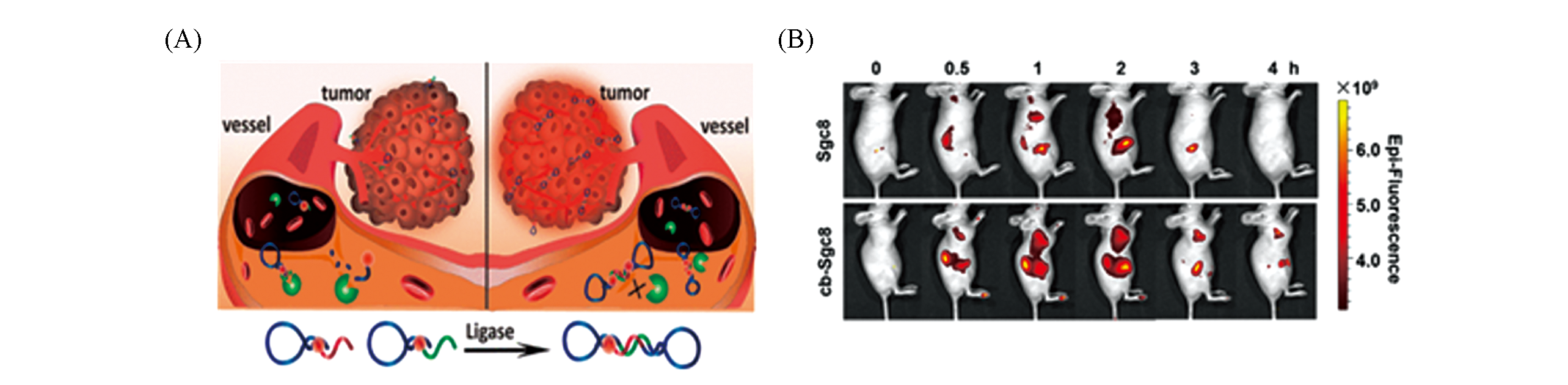
Fig.3 Imaging of CCRF?CEM tumor?bearing mice with sgc8 or cb?sgc8[72](A) Schematic illustration of developing circular bivalent aptamers; (B) in vivo fluorescence imaging of CCRF-CEM tumor-bearing mice after Cy5-labeled Sgc8 or cb-Sgc8 was injected through tail vein. Copyright 2017, American Chemical Society.
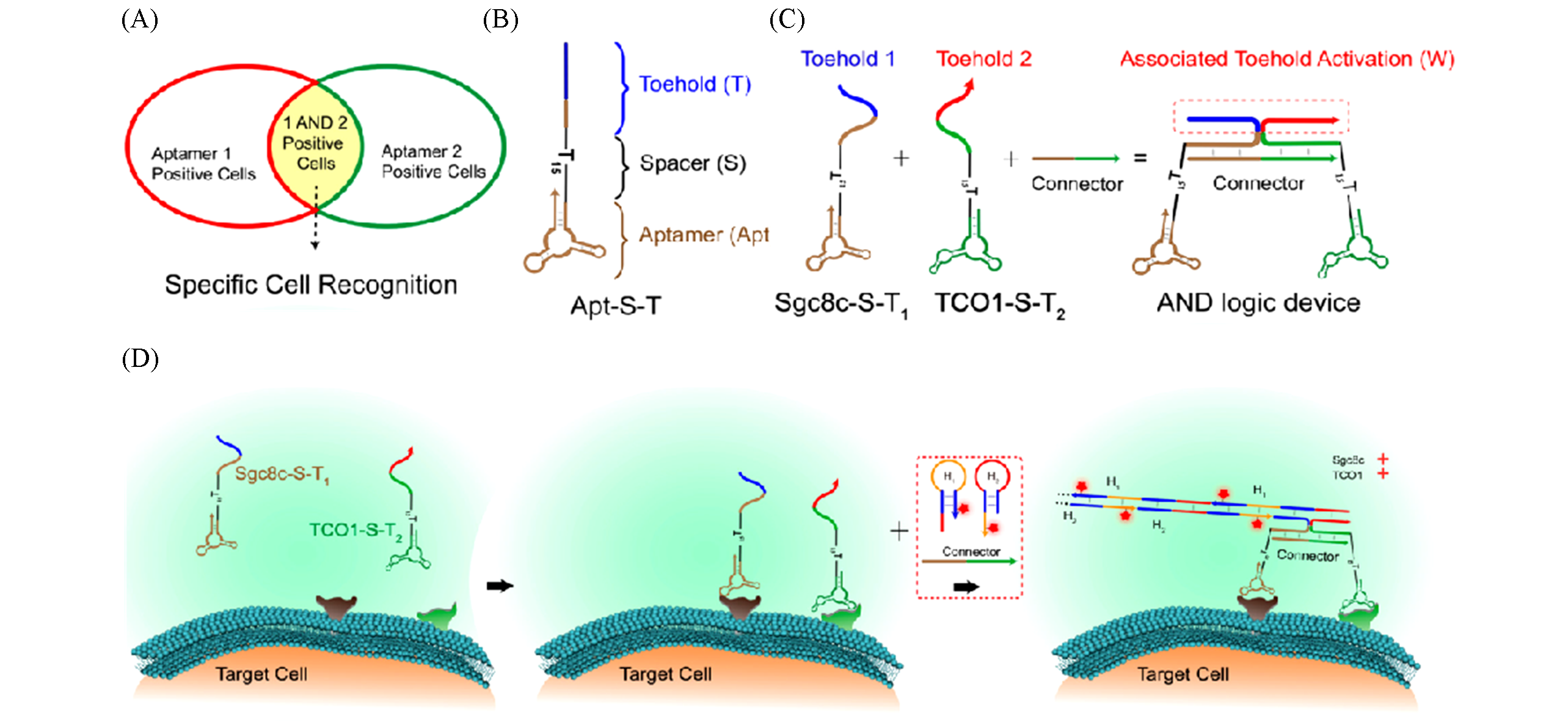
Fig.4 System design and operational mechanism[79](A) General principle of dual-aptamer-based AND logic device for cell identification and isolation; (B) structure and functional domains of aptamer-spacer-toehold(Apt-S-T) probe; (C) mechanism of dual-aptamer-based associative toehold activation. The Connector strand can bind with both Sgc8c-S-T1 and TCO1-S-T2 to form associated toeholds for HCR amplification reactions; (D) scheme of dual-aptamer-based AND logic device for target cell recognition and signal amplification.Copyright 2019, American Chemical Society.
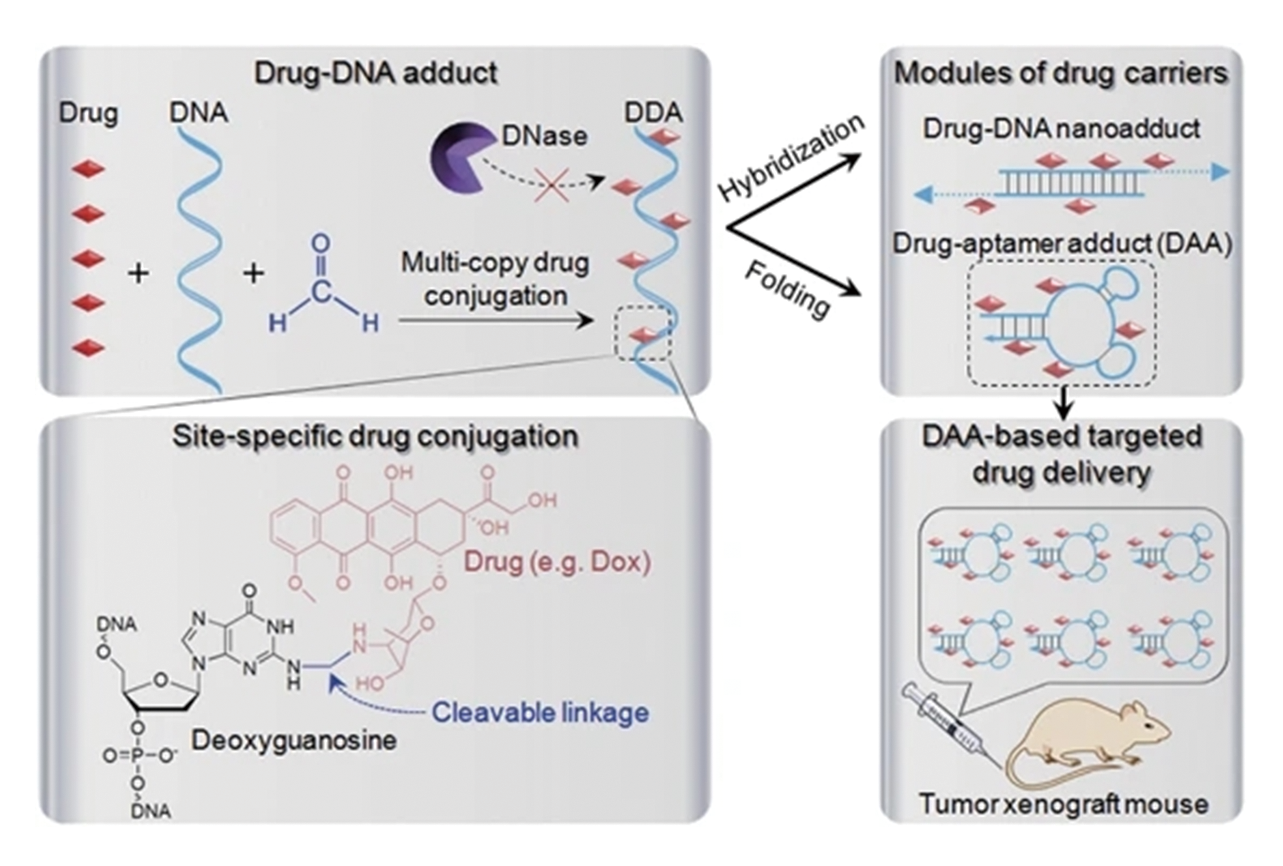
Fig.5 Schematic illustration of nuclease?resistant synthetic drug?DNA adducts as a simple, yet versatile and programmable platform for targeted anticancer drug delivery[100]Copyright 2015, Springer Nature.
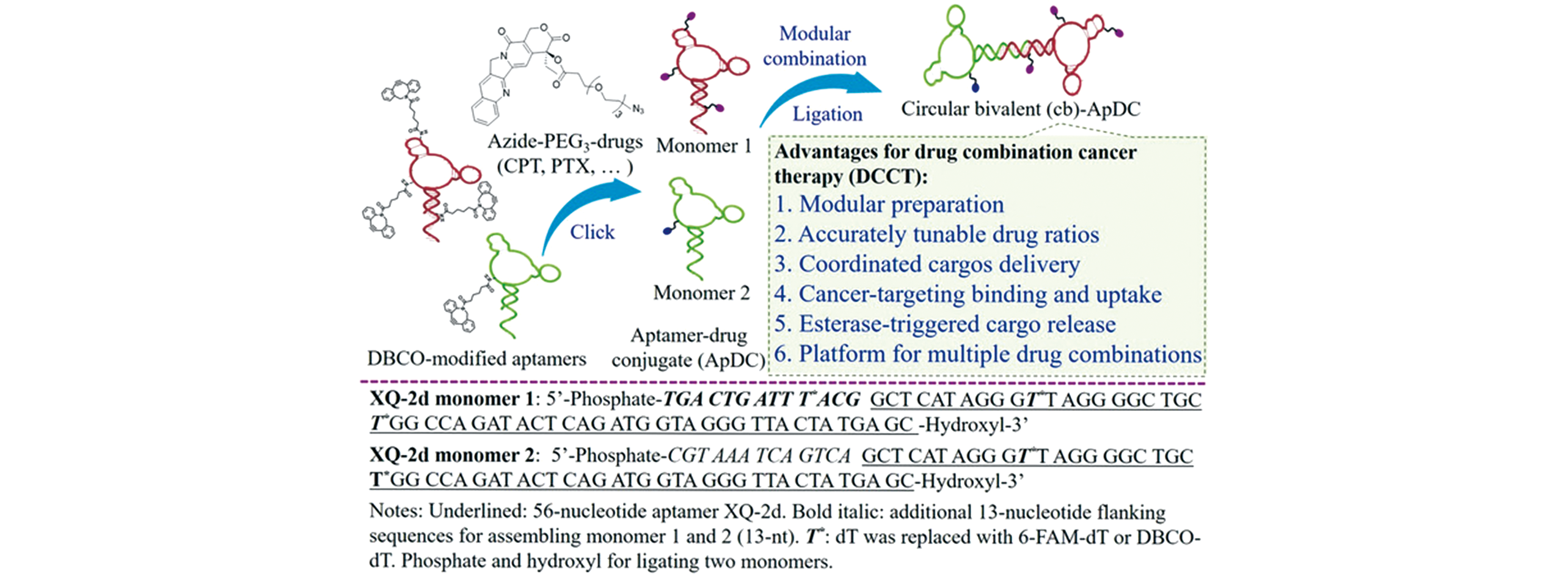
Fig.6 Preparation of circular bivalent aptamer?drug conjugates(cbApDCs) with accurate tunability of drug ratios for drug combination cancer therapy(DCCT) and the basic information of monomeric aptamers[102]Copyright 2019, Wiley-VCH.
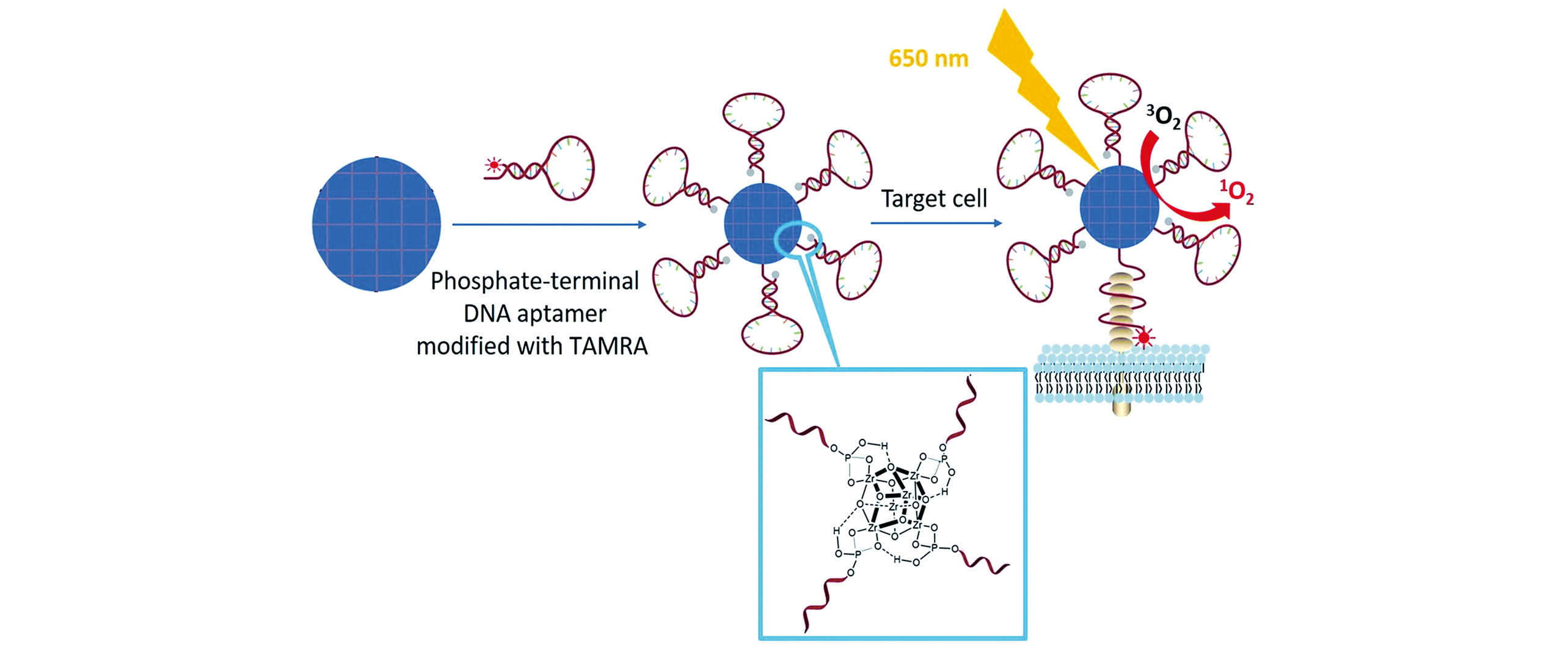
Fig.7 Illustration of phosphate?terminal DNA aptamer conjugation to a Zr?MOF nanoparticle quencher for target?induced imaging and photodynamic therapy[108]Copyright 2018, Royal Society of Chemistry.
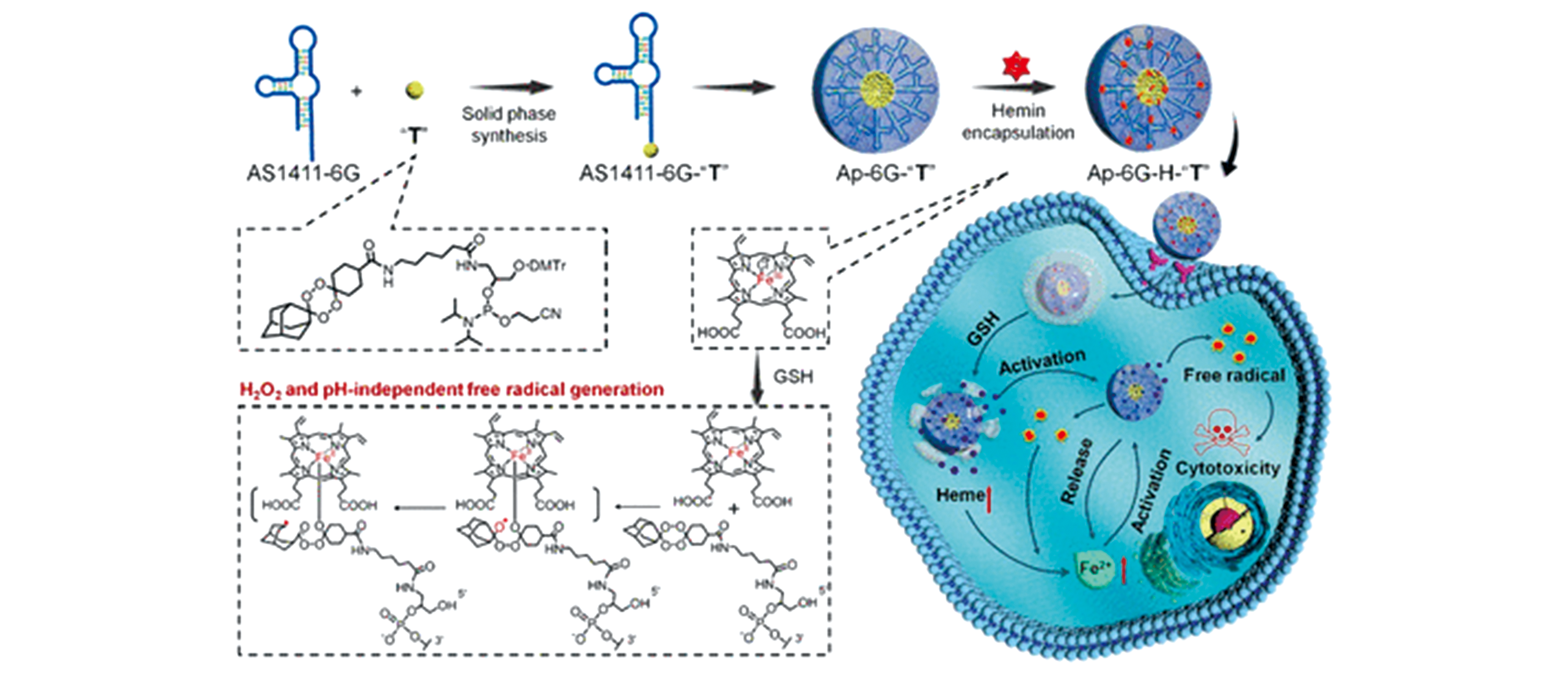
Fig.8 Schematic illustration of bioorthogonal ApPdC micelles for self?circulation and in?situ amplified generation of toxic free radical in cancer cells[114]Copyright 2020, American Chemical Society.
| 1 | Zeng H. M., Zheng R. S., Guo Y. M., Int. J. Cancer, 2015, 136(8), 1921—1930 |
| 2 | Zeng H. M., Chen W. Q., Zheng R. S., The Lancet. Global Health, 2018, 6(5), e555—e567 |
| 3 | Miller A. B., J. Public Health Pol., 1993, 14(4), 403—412 |
| 4 | Smith R. A., Cokkinides V., Eyre H. J., CA: Cancer J. Clin., 2000, 52(1), 8—22 |
| 5 | Swan J., Breen N., Lee N. C., Cancer, 2003, 97(6), 1528—1540 |
| 6 | Wiegering A., Ackermann S., Klein I., Int. J. Colorectal Dis., 2016, 31(5), 1039—1045 |
| 7 | Byers T., Wender R. C., Brawley O. W., CA: Cancer J. Clin., 2016, 66(5), 359—369 |
| 8 | Paziewska A., Mikula M., Ostrowski J., Prostate, 2018, 78(3), 178—185 |
| 9 | Farokhzad O. C., Karp J. M., Langer R. Expert Opin. Drug Del., 2006, 3(3), 311—324 |
| 10 | Farokhzad O. C., Cheng J. J., Langer R., Proc. Natl. Acad. Sci. USA, 2006, 103(16), 6315—6320 |
| 11 | McNamara J. O., Andrechek E. R., Giangrande P. H., Nat. Biotechnol., 2006, 24(8), 1005—1015 |
| 12 | Dhar S., Gu F. X., Lippard S. J., Proc. Natl. Acad. Sci. USA, 2008, 105(45), 17356—17361 |
| 13 | Xiong X. L., Liu H. P., Tan W. H., Angew. Chem. Int. Ed., 2013, 125(5), 1512—1516 |
| 14 | Dunn M., Jimenez R., Chaput J., Nat. Rev. Chem., 2017, 1, 0076 |
| 15 | Zhu G. Z., Chen X. Y., Adv. Drug Deliver Rev., 2018, 134, 65—78 |
| 16 | Alshaer W., Hillaireau H., Fattal E., Adv. Drug Deliver. Rev., 2018, 134, 122—137 |
| 17 | Chen C. H., Chernis G. A., Hoang V. Q., Proc. Natl. Acad. Sci. USA, 2003, 100, 9226—9231 |
| 18 | Ellington A. D., Szostak J. W., Nature, 1990, 346, 818—822 |
| 19 | Zhao Z. L., Fan H. H., Tan W. H., J. Am. Chem. Soc., 2014, 136, 11220 |
| 20 | Chen T., Shukoor M. I., Tan W. H., ACS Nano, 2011, 5, 7866 |
| 21 | Pu Y., Liu Z. X., Tan W. H., Anal. Chem.,2015, 87(3), 1919—1924 |
| 22 | Bai H. R., Fan H. H., Zhang X. B., Chen Z., Tan W. H., Acta Phys.⁃Chim. Sin., 2018, 34(4), 348—360(白华荣, 范换换, 张晓兵, 陈卓, 谭蔚泓. 物理化学学报, 2018, 34(4), 348—360) |
| 23 | Rothlisberger P., Hollenstein M., Adv. Drug Delivery Rev., 2018, 134, 3—21 |
| 24 | Nutiu R., Li Y. F., J. Am. Chem. Soc., 2003, 125(16), 4771—4778 |
| 25 | Famulok M., Mayer G., Acc. Chem. Res., 2011, 44(12), 1349—1358 |
| 26 | Tan W. H., Donovan M. J., Jiang J. H., Chem. Rev., 2013, 113(4), 2842—2862 |
| 27 | Sun H. G., Tan W. H., Zu Y. L., Analyst, 2016, 141, 403—415 |
| 28 | Dickey D. D., Giangrande P. H., Methods, 2016, 97, 94—103 |
| 29 | Yang X. H., Wang S. F., Wang K. M., Luo X. M., Tan W. H., Cui L., Chem. J. Chinese Universities, 2009, 30(5), 899—902(羊小海, 王胜锋, 王柯敏, 罗晓明, 谭蔚泓, 崔亮. 高等学校化学学报, 2009, 30(5), 899—902) |
| 30 | Shen Q. L., Xu L., Fang X. H., Adv. Mater., 2013, 25(16), 2368—2373 |
| 31 | Ma H. T., Liu J., Wan Y., Chem. Soc. Rev., 2015, 44, 1240—1256 |
| 32 | Song Y. L., Zhu Z., Yang C. Y., Anal. Chem., 2013, 85(8), 4141—4149 |
| 33 | Hassan E. M., Willmore W. G., DeRosa M. C., Nucleic Acid. Ther., 2016, 26(6), 335—347 |
| 34 | Keefe A., Pai S., Ellington A., Nat. Rev. Drug Discov., 2010, 9, 537—550 |
| 35 | Ireson C. R., Kelland L. R., Mol. Cancer Ther., 2016, 5(12), 2957-2962 |
| 36 | Fu Z. Y., Xiang J., Int. J. Mol. Sci., 2020, 21, 2793 |
| 37 | Cerchia L., Cancers, 2018, 10(5), 132—136 |
| 38 | Peng T. H., Deng Z. Y., Tan W. H., Coordin. Chem. Rev., 2020, 403, 213080 |
| 39 | Kim D. K., Jeong Y. Y., Jon S. Y., ACS Nano, 2010, 4(7), 3689—3696 |
| 40 | Medley C. D., Bamrungsap S., Tan W. H., Anal. Chem., 2011, 83(3), 727—734 |
| 41 | Lao Y. H., Phua K. K., Leong K. W., ACS Nano, 2015, 9(3), 2235—2254 |
| 42 | Zhu G. Z., Niu G., Chen X. Y., Bioconjugate Chem., 2015, 26(11), 2186—2197 |
| 43 | Meng H. M., Fu T., Tan W. H., Natl. Sci. Rev., 2015, 2(1), 71—84 |
| 44 | Tan X. Y., Jia F., Zhang K., J. Controled Release, 2020, 323, 240—252 |
| 45 | Poolsup S., Kim C. Y., Curr. Opin. Biotech., 2017, 48, 180—186 |
| 46 | Osborne S. E., Ellington A. D., Chem. Rev., 1997, 97(2), 349—370 |
| 47 | Nutiu R., Li Y. F., Angew. Chem. Int. Ed., 2005, 44(7), 1061—1065 |
| 48 | Wilson D. S., Szostak J. W., Annu. Rev. Biochem., 1999, 68(1), 611—647 |
| 49 | Fang X. H., Tan W. H., Acc. Chem. Res., 2010, 43, 1, 48—57 |
| 50 | Ellington A. D., Szostak J. W., Nature, 1990, 346, 818—822 |
| 51 | Tuerk C., Gold L., Science, 1990, 249, 505—510 |
| 52 | Sefah K., Meng L., Tan W. H., PLoS One, 2010, 5(12), e14269 |
| 53 | Shangguan D. H., Li Y., Tan W. H., Proc. Nat. Acad. Sci., 2006, 103(32), 11838—11843 |
| 54 | Jin C., Zheng J., Tan W. H., J. Mol. Evol., 2015, 81, 162—171 |
| 55 | Sefah K., Shangguan D. H., Tan W. H., Nat. Protoc., 2010, 5(6), 1169 |
| 56 | Zhu G. Z., Ye M., Tan W. H., Chem. Commun., 2012, 48(85), 10472—10480 |
| 57 | Kunii T., Ogura S., Kobatake E., Analyst, 2011, 136, 1310—1312 |
| 58 | Sefah K., Tang Z. W., Tan W. H., Leukemia, 2009, 23, 235—244 |
| 59 | Wu X. Q., Zhao Z. L., Tan W. H., Theranostics, 2015, 5(9), 985—994 |
| 60 | Zhao Z. L., Xu L., Tan W. H., Analyst, 2009, 134(9), 1808—1814 |
| 61 | Hicke B. J., Stephens A. W., Schmidt P. G., J. Nucl. Med., 2006,47(4), 668—678 |
| 62 | Moosavian S. A., Jaafari M. R., Abnous K., Iran. J. Basic Med. Sci., 2015, 18(6), 576—586 |
| 63 | Famulok M., Hartig J. S., Mayer G., Chem. Rev., 2007, 107(9), 3715—3743 |
| 64 | Ni S. J, Yao H. Z., Zhang G., Int. J. Mol. Sci., 2017, 18(8), 1683—1704 |
| 65 | Ganson N. J., Povsic T. J., Hershfifield M. S., Allergy Clin. Immunol., 2016, 137, 1610—1613 |
| 66 | Morita Y., Leslie M., Tanaka T., Cancers, 2018, 10(3), 80—102 |
| 67 | Jaschke A., Furste J. P., Erdmann V. A., Nucleic. Acids. Res., 1994, 22(22), 4810—4817 |
| 68 | Kartschem C., Levy M., Nucleic Acid. Ther., 2017, 27(6), 335—344 |
| 69 | Li S., Xu H., Shao N. S., J. Pathol., 2009, 218(1), 327—336 |
| 70 | Li J. L., Zheng H. Y., Bates P. J., Malik T.,Li X. F., Trent J. O., Ng C. K., Nucl. Med. Biol., 2014, 41(2), 179—185 |
| 71 | Wang S., Kong H., Gong X. Y., Zhang S. C., Zhang X. R., Anal. Chem., 2014, 86(16), 8261-8266 |
| 72 | Kuai H. L., Zhao Z. L., Tan W. H., J. Am. Chem. Soc., 2017, 139(27), 9128—9131 |
| 73 | Zhong W., Pu Y., Liu H. X., Anal. Chem., 2019, 91(13), 8289—8297 |
| 74 | Zou Y. X., Huang S. Q., Liao Y. X., Zhu X. P., Chen Y. Q., Chen L., Liu F., Hu X. X., Tu H. J., Zhang L., Liu Z. K., Chen Z., Tan W. H., Chem. Sci., 2018, 9(10), 2842—2849 |
| 75 | Jiang Y., Shi M. L, Tan W. H., Angew. Chem. Int. Ed., 2017, 56(39), 11916—11920 |
| 76 | Wang S., Zhang L. Q., Tan W. H., ACS Nano, 2017, 11(4), 3943—3949 |
| 77 | Wu Y. R., Sefah K., Tan W. H., Proc. Nat. Acad. Sci., 2010, 107(1), 5—10 |
| 78 | Song Y. L., Shi Y. Z., Yang C. Y., Angew. Chem. Int. Ed., 2019, 58(8), 2236—2240 |
| 79 | Chang X., Zhang C., Hao D., Tan W. H., J. Am. Chem. Soc., 2019, 141(32), 12738—12743 |
| 80 | Ruckman J., Green L. S., Janjic N., J. Biol. Chem., 1998, 273(32), 20556—20567 |
| 81 | Li N., Nguyen H. H., Ellington A. D., Chem. Commun., 2010, 46(3), 392—394 |
| 82 | Ismail S. I., Alshaer W., Adv. Drug. Deliver. Rev., 2018, 134, 51—64 |
| 83 | Dollins C. M., Nair S., Sullenge B. A., Chem. Biol., 2008, 15(7), 675—682 |
| 84 | Parekh P., Kamble S., Zu Y. L., Biomaterials, 2013, 34(35), 8909—8917 |
| 85 | Mahlknecht G., Maron R., Yarden Y., Proc. Natl. Acad. Sci. USA, 2013, 110(20), 8170—8175 |
| 86 | McNamara J. O., Kolonias D., Gilboa E., J. Clin. Invest., 2008, 118(1), 376—386 |
| 87 | Teller C., Willner I., Curr. Opin. Biotechnol., 2010, 21(4), 376—391 |
| 88 | Zhou J. H., Rossi J., Nat. Rev. Drug Discov., 2017, 16(3), 181—202 |
| 89 | Ng E. W. M., Shima D. T., Adamis A. P., Nat. Rev. Drug Discov., 2006, 5(2), 123—132 |
| 90 | Yazdian⁃Robati R., Bayat P., Abnous K., Int. J. Biol. Macromol., 2020, 155(15), 1420—1431 |
| 91 | Hoellenriegel J., Zboralski D., Burger J. A., Blood, 2014, 123(7), 1032—1039 |
| 92 | Guo F., Wang Y., Zhang W., Oncogene, 2016, 35(7), 816—826 |
| 93 | Huang Y. F., Shangguan D. H., Tan W. H., ChemBioChem, 2009, 10(5),862—868 |
| 94 | Bagalkot V., Farokhzad O. C., Jon S. Y., Angew. Chem. Int. Ed., 2006, 45(48), 8149—8152 |
| 95 | Chen K., Liu B., Tan W. H., Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol., 2017, 9(3), e1438—e1450 |
| 96 | Xuan W. J., Peng Y. B., Tan W. H., Biomaterials, 2018, 182, 216—226 |
| 97 | Trinh T. L., Zhu G. Z., Tan W. H., PLoS One, 2015, 10(11), e0136673 |
| 98 | He J. X., Peng T. H., Tan W. H., J. Am. Chem. Soc., 2020, 142(6), 2699—2703 |
| 99 | Yang Q. X., Deng Z. Y., Tan W. H., J. Am. Chem. Soc., 2020, 142(5), 2532—2540 |
| 100 | Zhu G. Z., Cansiz S., Tan W. H., Npg Asia Materials, 2015, 7(3), e169—e177 |
| 101 | Deng Z. Y., Yang Q. X., Tan W. H., Bioconjugate Chem., 2020, 31(1), 37—42 |
| 102 | Zhou F., Wang P., Tan W. H., Angew. Chem. Int. Ed., 2019, 131(34), 11787—11791 |
| 103 | Lv C., Yang C., Hao D., Tan W. H., Anal. Chem., 2019, 91(21), 13818—13823 |
| 104 | Qiu L. P., Chen T., Tan W. H., Nano Letters, 2015, 15(1), 457—463 |
| 105 | Niu W. J., Chen X. G., Tan W. H., Veige A. S., Angew. Chem. Int. Ed., 2016, 55(31), 8889—8893 |
| 106 | Niu W. J., Teng I., Tan W. H., Veige A. S., Dalton Trans.,2018, 47(1), 120—126 |
| 107 | Wang X. W., Gao W., Tan W. H., Nanoscale, 2016, 8, 7942—7948 |
| 108 | Liu Y., Hou W. J., Tan W. H., Chem. Sci., 2018, 9, 7505—7509 |
| 109 | Li J., Wu S. X., Tan W. H., Nanoscale, 2016, 8(16), 8600—8606 |
| 110 | Setyawati M. I., Tay C. Y., Leong D. T., ACS Nano, 2017, 11(5), 5020—5030 |
| 111 | Peng F., Setyawati M. I., Leong D. T., Nat. Nanotechnol., 2019, 14(3), 279—286 |
| 112 | Zou J. M., Shi M. L., Tan W. H., Angew. Chem. Int. Ed., 2019, 91(3), 2425—2430 |
| 113 | Yang L., Sun H., Tan W. H., Angew. Chem. Int. Ed., 2018, 130(52), 17294—17298 |
| 114 | Xuan W. J., Xia Y. H., Tan W. H., J. Am. Chem. Soc., 2019, 142(2), 937—944 |
| [1] | 刘珂, 靳宇, 梁建功, 吴园. 化学修饰提高核酸适体结合亲和力的研究进展[J]. 高等学校化学学报, 2021, 42(11): 3477. |
| [2] | 张晓荣, 陈岚岚, 胡善文. 基于分子识别的细菌检测研究进展[J]. 高等学校化学学报, 2021, 42(11): 3468. |
| [3] | 吉采灵, 程兴, 谈洁, 袁荃. 功能化核酸适体的筛选及分子识别应用[J]. 高等学校化学学报, 2021, 42(11): 3457. |
| [4] | 黄玲, 庄梓健, 李翔, 石沐玲, 刘高强. 基于核酸适体的外泌体分子识别研究进展[J]. 高等学校化学学报, 2021, 42(11): 3493. |
| [5] | 解忱, 陈娜, 杨雁冰, 袁荃. 核酸适体功能化的二维材料场效应晶体管传感器研究进展[J]. 高等学校化学学报, 2021, 42(11): 3406. |
| [6] | 赵卓, 王雪强. 核酸适体偶联药物的生物偶联构建技术与应用[J]. 高等学校化学学报, 2021, 42(11): 3367. |
| [7] | 刘学娇, 杨帆, 刘爽, 张春娟, 刘巧玲. 核酸适体靶向的膜蛋白识别与功能调控研究进展[J]. 高等学校化学学报, 2021, 42(11): 3277. |
| [8] | 赵秋伶, 刘玲玲, 杨丽娜. 基于核酸适体检测ATP的酶联分析新方法[J]. 高等学校化学学报, 2014, 35(6): 1161. |
| [9] | 吴尚荣, 金冰, 张楠, 柳影, 刘祥军, 李松青, 上官棣华. 一种不对称菁染料及其与不同结构DNA的相互作用[J]. 高等学校化学学报, 2014, 35(10): 2085. |
| [10] | 赵春玲, 华梅, 赵晓慧, 黄亮亮, 冯亚娟, 牛司朋, 符雪文, 袁聪, 杨云慧. 非标记型血小板衍生生长因子核酸适体传感器[J]. 高等学校化学学报, 2013, 34(1): 61. |
| [11] | 何微娜, 涂熹娟, 郭祥群. 牛血清白蛋白修饰的近红外荧光纳米金顺磁标记分子探针[J]. 高等学校化学学报, 2012, 33(11): 2411. |
| [12] | 黄艳琴, 秦伟胜, 任厚基, 曹国益, 刘兴奋, 黄维. 基于阳离子型共轭聚合物和核酸适体的腺苷检测新方法[J]. 高等学校化学学报, 2012, 33(10): 2213. |
| [13] | 刘雪平, 欧阳湘元, 吴会旺, 沈国励, 俞汝勤. 基于目标物诱导置换及纳米金催化的新型荧光技术检测腺苷[J]. 高等学校化学学报, 2012, 33(08): 1692. |
| [14] | 郑静, 秦佳华, 龚晨, 黄焕, 何品刚, 方禹之. 基于核酸适体与互补核酸和目标蛋白之间竞争反应的PDGF蛋白特异性识别[J]. 高等学校化学学报, 2011, 32(11): 2509. |
| [15] | 郭秋平, 羊小海, 王柯敏, 孟祥贤, 李军, 仲志鸿. 双链荧光核酸适体探针用于蛋白质的简便快速检测[J]. 高等学校化学学报, 2010, 31(2): 274. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||