

高等学校化学学报 ›› 2025, Vol. 46 ›› Issue (1): 20240260.doi: 10.7503/cjcu20240260
• 综合评述 • 上一篇
收稿日期:2024-05-28
出版日期:2025-01-10
发布日期:2024-08-12
通讯作者:
代云路
E-mail:beili@um.edu.mo;yldai@um.edu.mo
作者简介:李 蓓, 女, 博士, 研究助理教授, 主要从事精准肿瘤治疗和免疫助剂开发等方面的研究. E-mail: beili@um.edu.mo
基金资助:
YAN Ziliang, LI Bei( ), DAI Yunlu(
), DAI Yunlu( )
)
Received:2024-05-28
Online:2025-01-10
Published:2024-08-12
Contact:
DAI Yunlu
E-mail:beili@um.edu.mo;yldai@um.edu.mo
Supported by:摘要:
超分子纳米药物递送平台因具有性质多样化、 药物释放可控及制备简易等特点而备受关注. 据报道, 富含酚羟基结构的多酚可与不同性质类药物产生非共价相互作用, 自组装成超分子纳米系统, 从而实现不同路径的药物递送. 同时, 多酚自身具备抗肿瘤、 抗菌、 抗氧化、 抗炎和保护心脏等功能, 这使得基于多酚的递送系统在疾病治疗方面前景广阔. 本综合评述概述了多酚型超分子纳米递送系统构建中包括的超分子相互作用力. 根据所负载药物的性质(例如疏水性药物、 蛋白质、 DNA等), 分类阐述了不同相互作用力在药物负载中发挥的功能. 最后, 对当前基于多酚的超分子纳米系统中存在的争议性问题进行了评述总结. 本文可为各种新兴的多酚基材料的设计和基础研究提供参考.
中图分类号:
TrendMD:
严子谅, 李蓓, 代云路. 基于多酚的超分子纳米药物递送系统的研究进展. 高等学校化学学报, 2025, 46(1): 20240260.
YAN Ziliang, LI Bei, DAI Yunlu. Research Progress in Supramolecular Drug Delivery Nanosystems Based on Polyphenols. Chem. J. Chinese Universities, 2025, 46(1): 20240260.
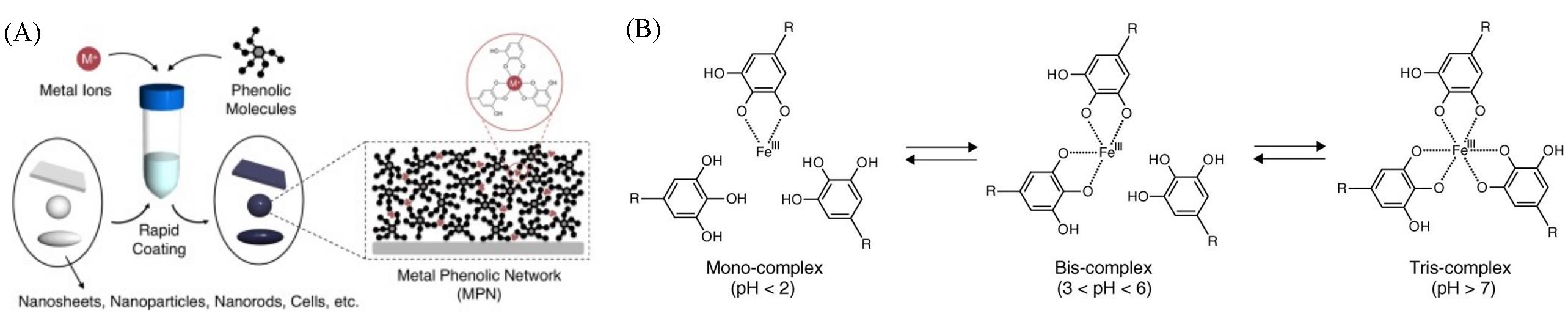
Fig.2 Basic construction method(A) and its coordination mechanism(B) of metal⁃polyphenol supramolecular network[12]Copyright 2013, American Association for the Advancement of Science.
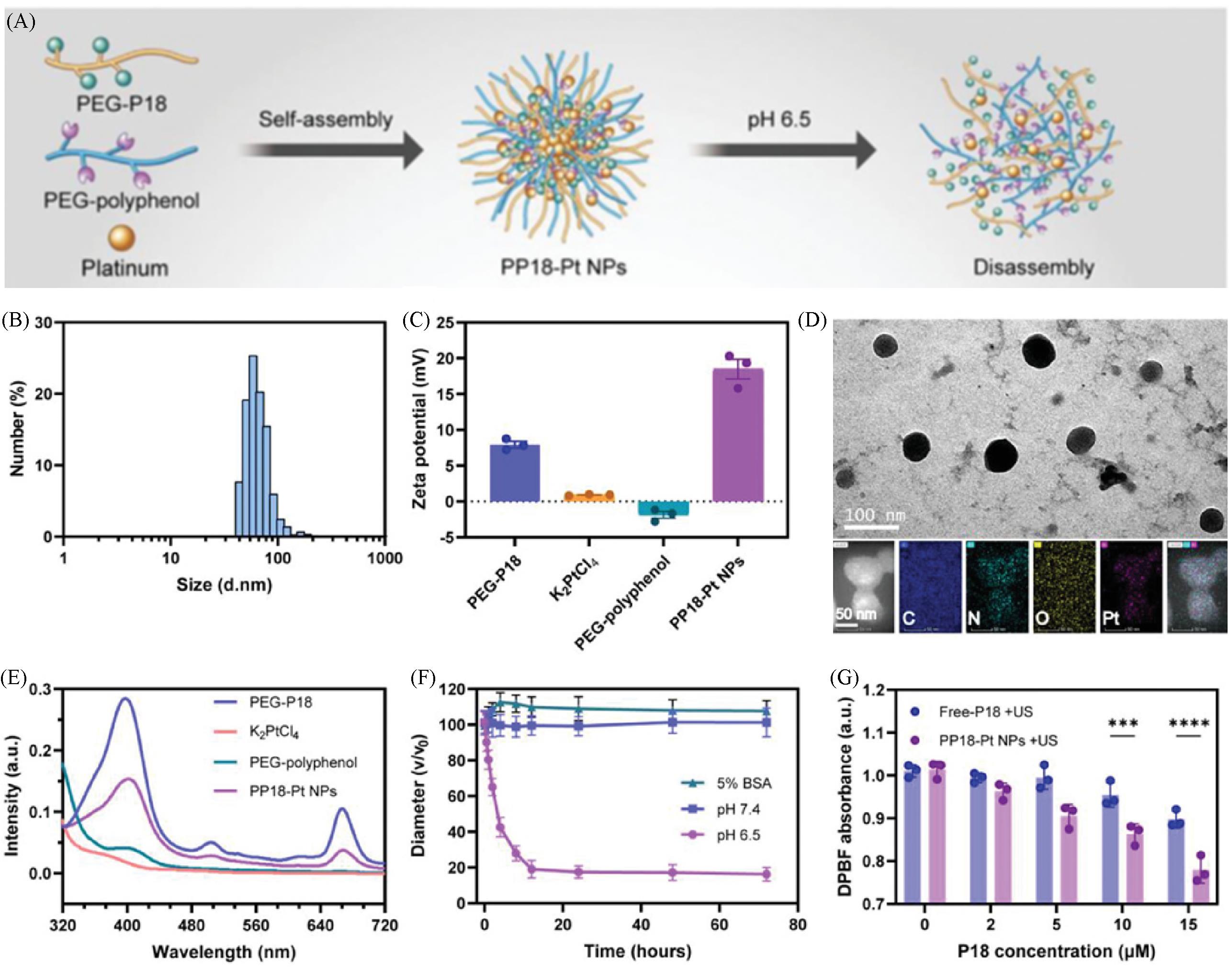
Fig.5 Schematic assembly⁃disassembly process(A), size distribution(B), zeta potential(C), TEM image(D), UV⁃Vis spectrum(E), stability of hydrated size(F) and ROS generation(G) of PP18⁃Pt NPs[35]Copyright 2022, John Wiley and Sons.
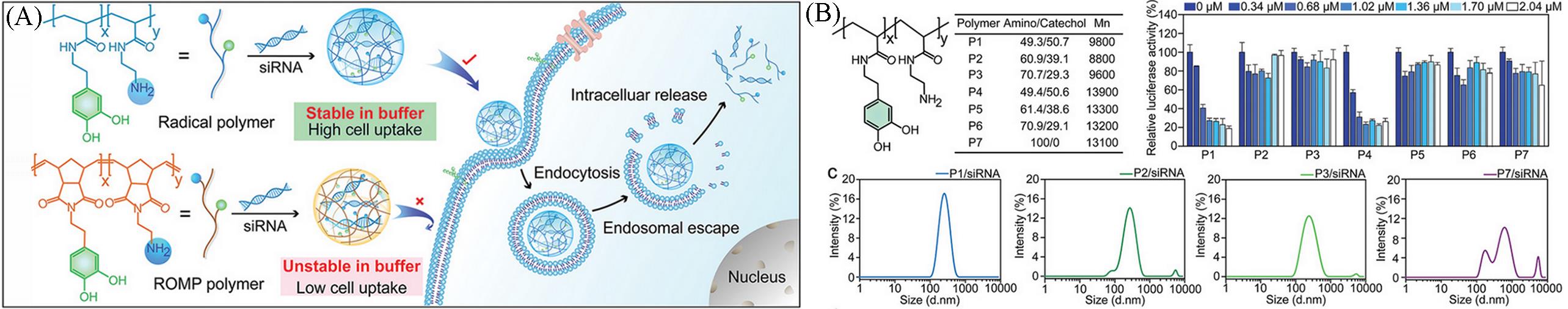
Fig.7 Intracellular siRNA delivery procedure(A) and siRNA⁃mediated luciferase gene knockdown efficiency(B) of cationic polyphenolic polymerin HeLa⁃Luc cells[45]Copyright 2021, John Wiley and Sons.
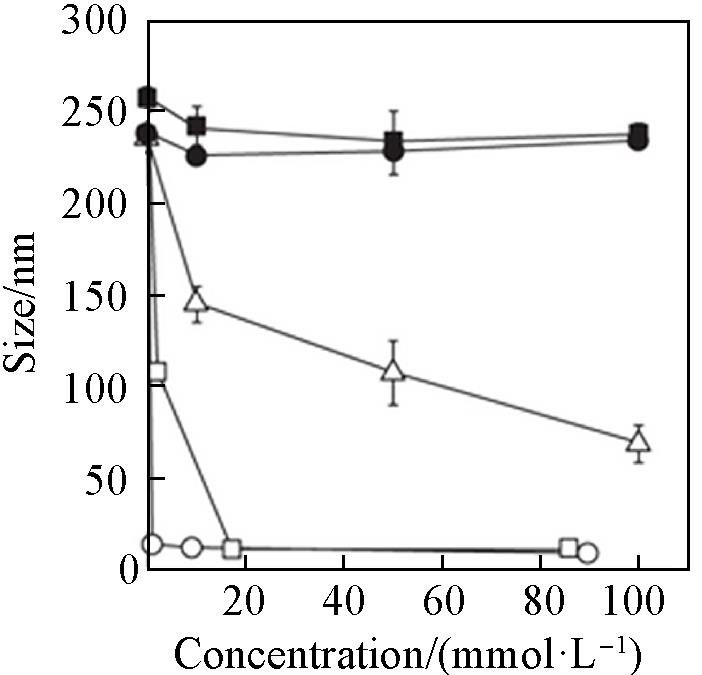
Fig.8 Particle size variations of EGCG⁃protein nanoparticles affected by different noncovalent interactions competitors including Tween 20(open circles), Triton X⁃100(open squares), SDS(open triangles), urea(filled circles) and NaCl(filled squares)[49]Copyright 2014, Springer Nature.
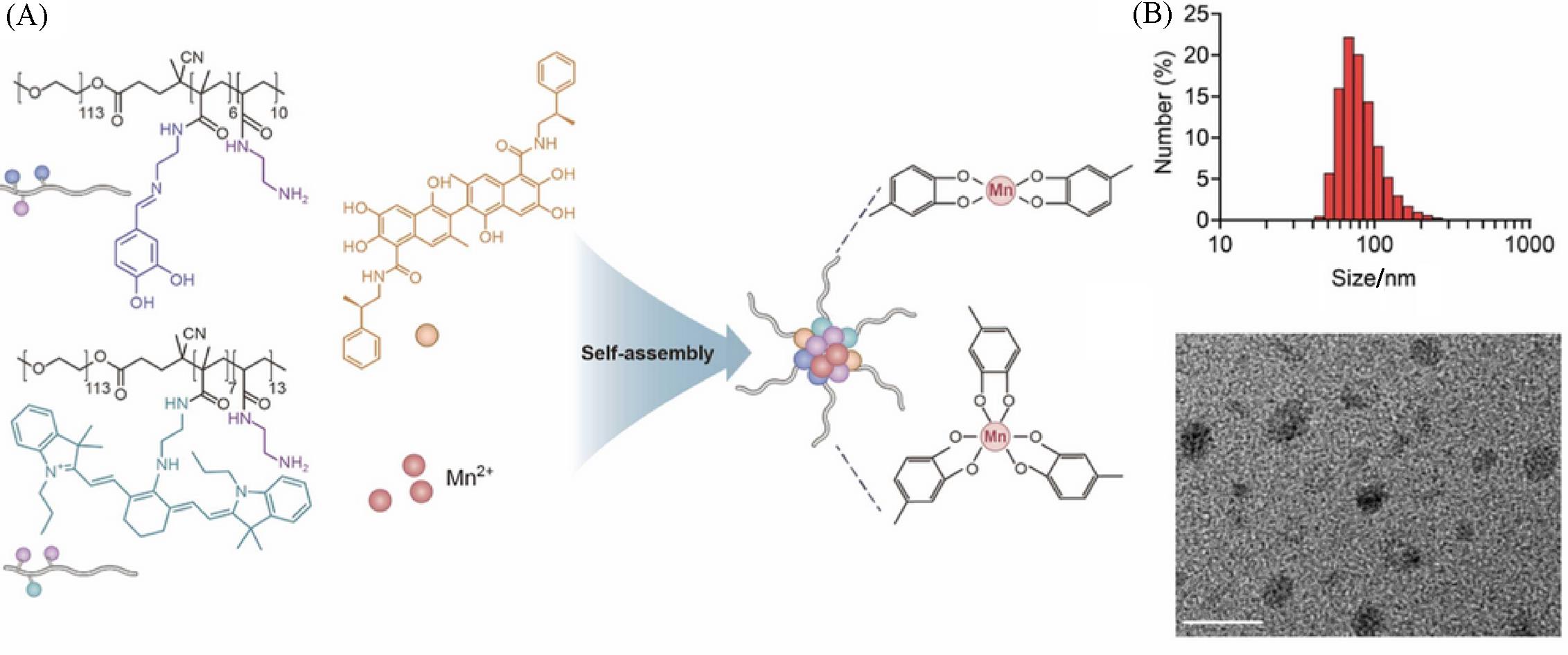
Fig.10 Nanoparticle formation(A), size distribution and TEM image(B) of amphiphilic polyphenolic polymer(PEG⁃Pho) encapsulating hydrophobic drugs and coordinating with Mn2+[52]Copyright 2022, Elsevier.
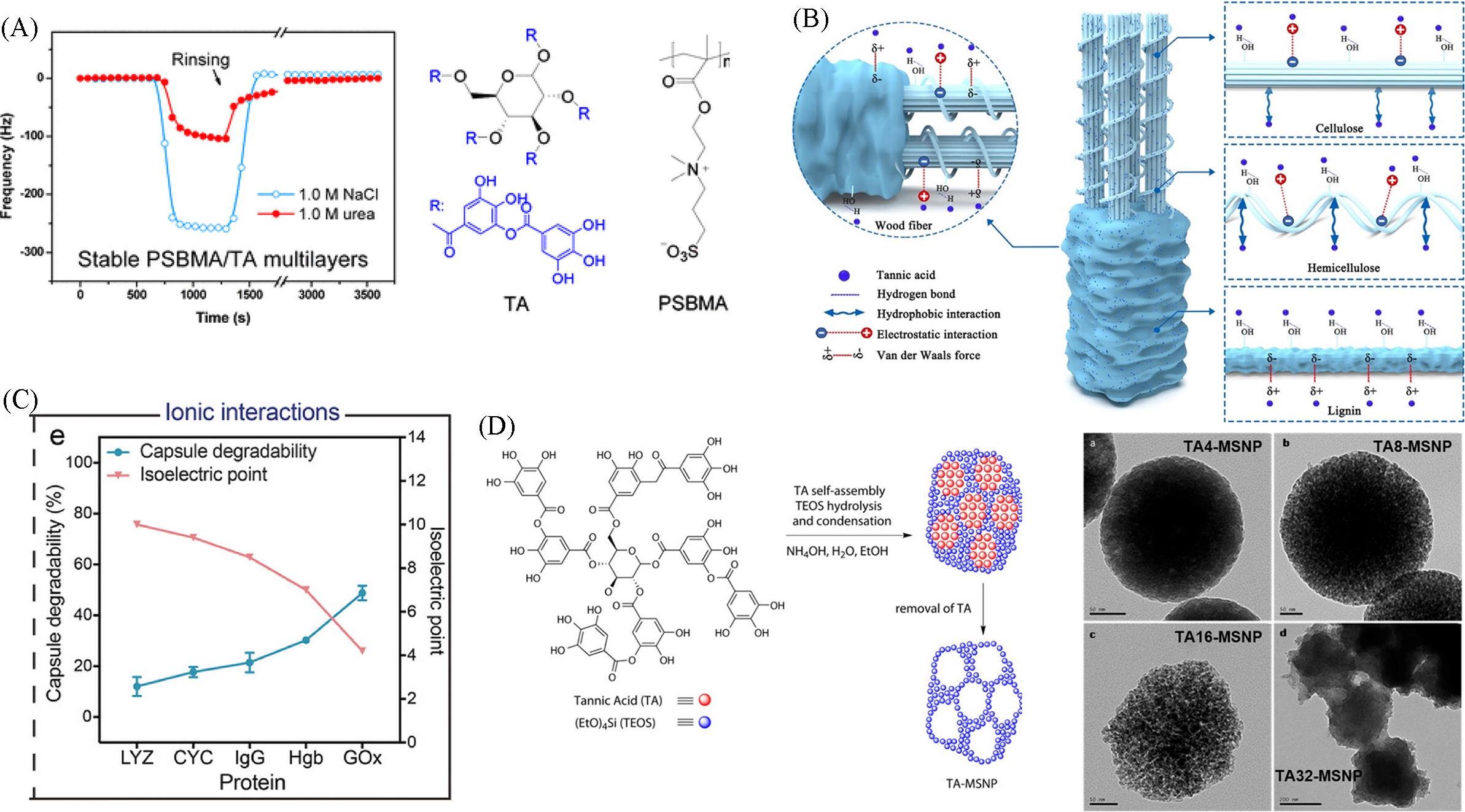
Fig.11 Stability of PSBMA/TA multilayers(A)[56], interaction between TA and wood fiber, cellulose, hemicellulose and lignin(B)[57], degradability of protein⁃polyphenol capsule(C)[50], construction and TEM image of TA⁃MSNP nanoparticles(D)[58](A) Copyright 2015, American Chemical Society; (B) Copyright 2024, American Chemical Society; (C) Copyright 2022, American Chemical Society; (D) Copyright 2014, American Chemical Society.
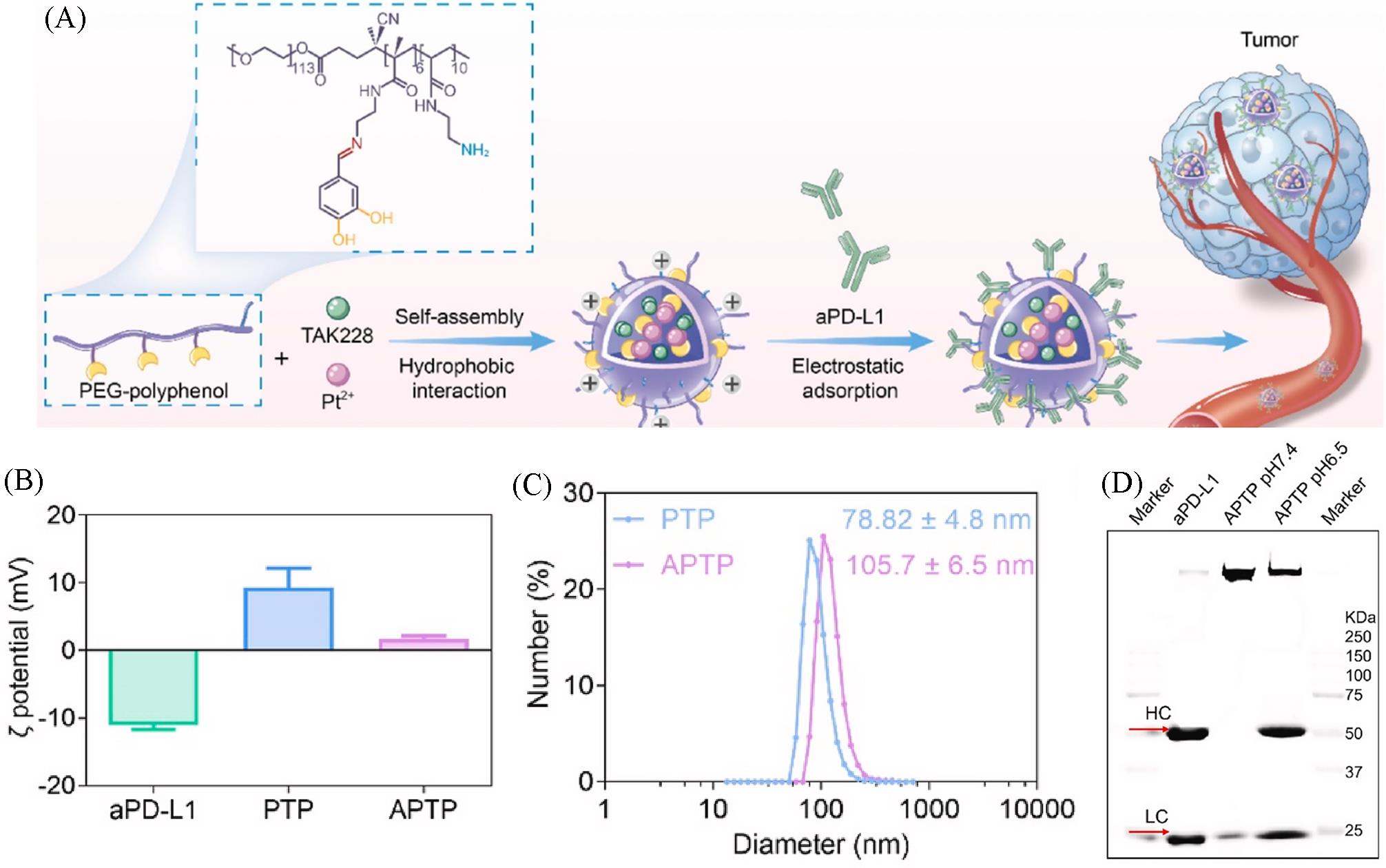
Fig.12 Synthetic procedure(A), zeta potential(B), particle size distribution(C) and aPD⁃L1 loading evaluation(D) of antibody⁃loaded PTP(APTP)[59]Copyright 2023, Elsevier.
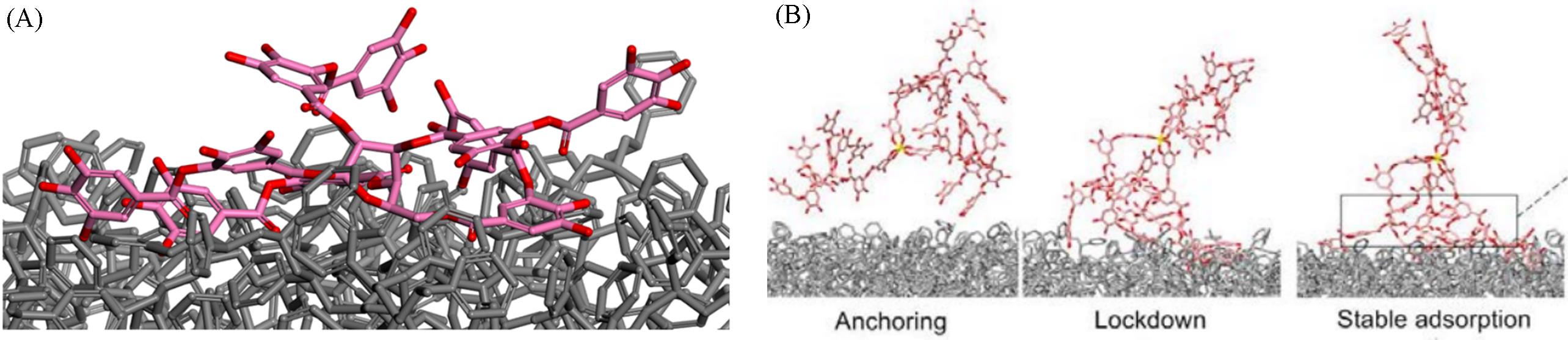
Fig.13 Typical equilibrium adsorption configuration(A) and representative snapshots obtained from molecular dynamics simulations showing the anchoring, locking, and stabilizing adsorption steps(B) between gallic groups of TA3/FeIII and the PS surface[14]Copyright 2016, Springer Nature.
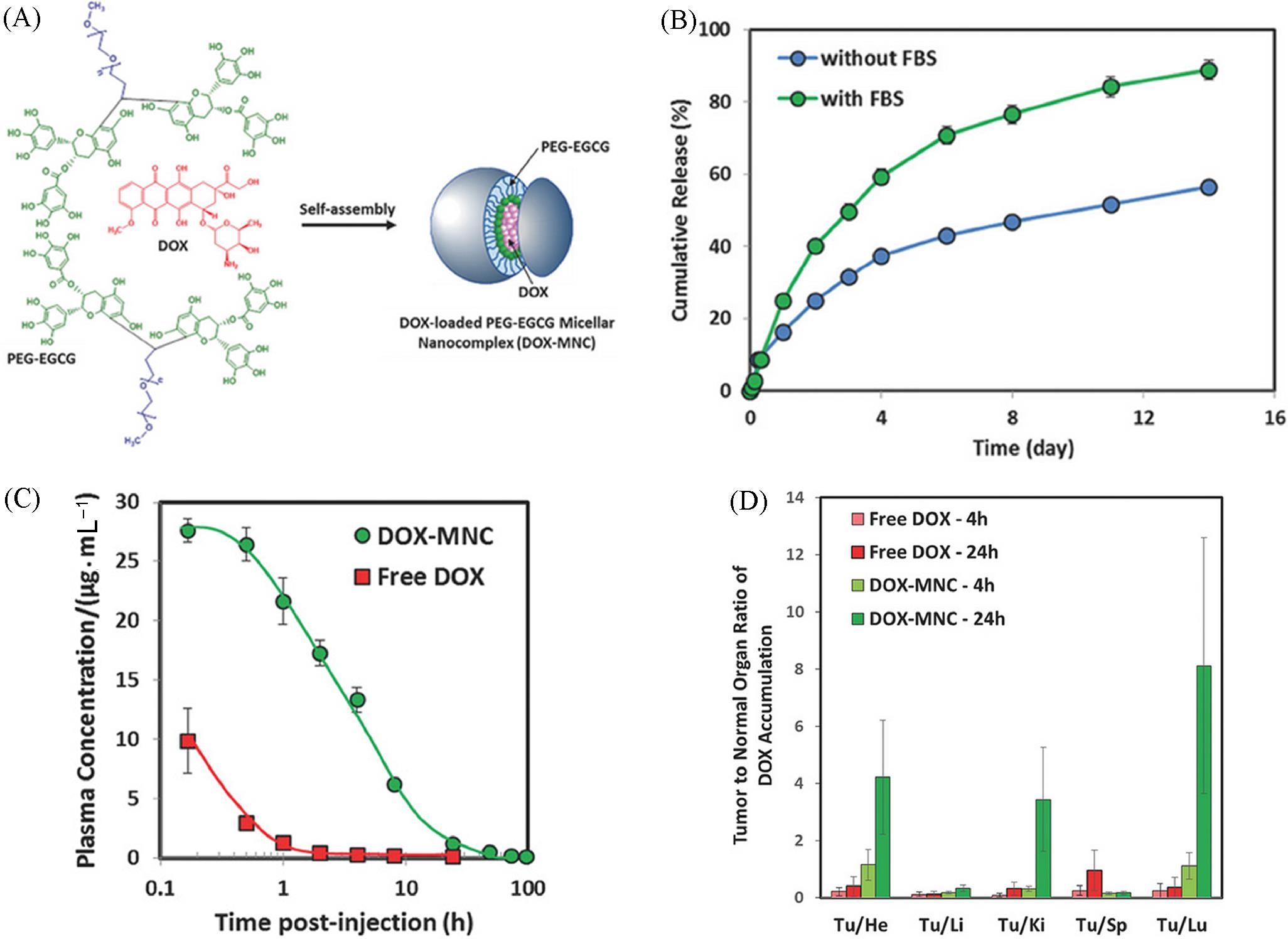
Fig.14 Assembly between PEG⁃EGCG and DOX based on π⁃π interaction to form DOX⁃MNC(A) and the corresponding DOX release in FBS(B), pharmacokinetic curve(C) as well as tumor accumulation(D)[64]Copyright 2018, John Wiley and Sons.
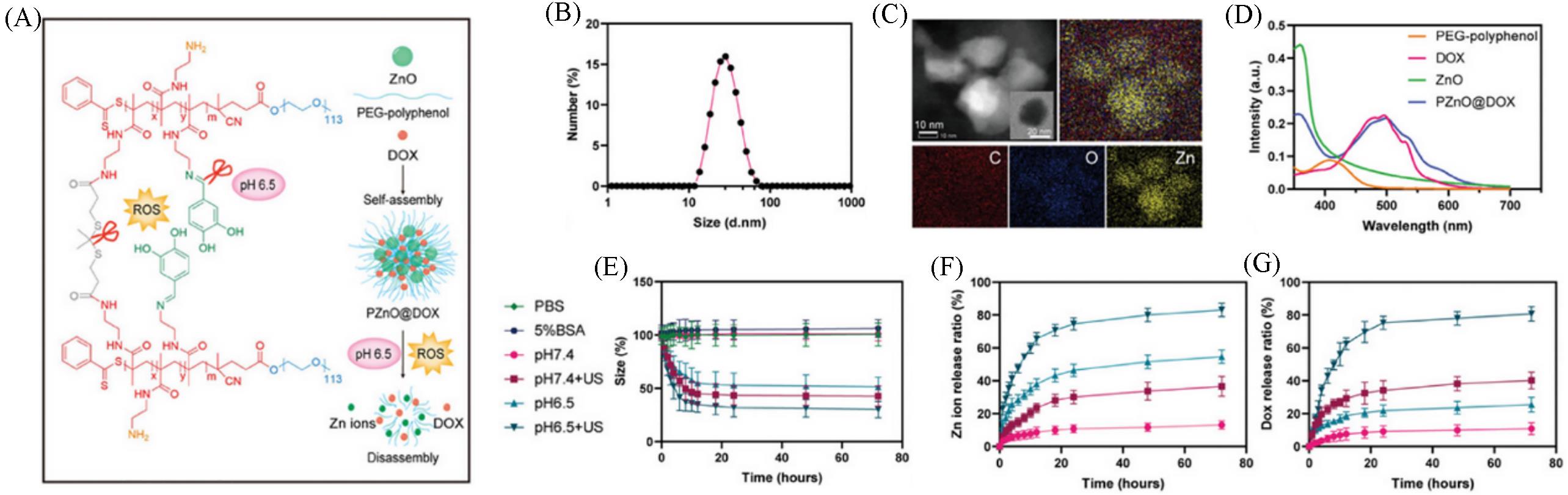
Fig.15 Preparation(A), size(B), morpgology(C), DOX loading(D) and drug release(E—G) of acid and ROS dual⁃responsive nanogel under different conditions including PBS(green rhombus), 5% BSA(hexagons), pH 7.4(pink circles), pH 7.4+US(purple squares), pH 6.5(triangles) and pH 6.5+US(inverted triangle)[18]Copyright 2024, John Wiley and Sons.
| Types of polyphenols | Loaded substance | Supramolecular interaction | Application | Ref. |
|---|---|---|---|---|
| Catechol | DOX, Zn2+ | Hydrophobic interactions, Coordination interactions | Tumor treatment | [ |
| EGCG, Catechol | W6+ | Hydrophobic interactions, Coordination interactions | Tumor treatment | [ |
| Gallic acid, Catechol | Hf4+, Hemoglobin | Hydrophobic interactions, Coordination interactions | Tumor treatment | [ |
| Rosmarinic acid | Dexamethasone | Hydrophobic interactions | Colitis | [ |
| Quercetin | None | Hydrophobic interactions, Hydrogen Bond | Colitis | [ |
| TA | Green fluorescent protein | Hydrophobic interactions | Heart disease | [ |
| Salvianolic acid B, Catechol | Ca2+ | Hydrophobic interactions, Coordination interactions | Renal fibrosis | [ |
| EGCG | Fe3+, DOX | Hydrophobic interactions, Coordination interactions, π⁃π interactions | Tumor treatment | [ |
| Polydopamine | Zn2+, DOX | Coordination interactions | Tumor treatment | [ |
| TA | BSA | Hydrophobic interactions, Hydrogen Bond | Colitis | [ |
| Catechol | 5⁃Aza, W6+ | Hydrophobic interactions, Coordination interactions | Tumor treatment | [ |
| TA | DOX | Hydrophobic interactions, Hydrogen Bond | Tumor treatment | [ |
| EGCG | DOX, Fe3+ | Hydrophobic interactions, Coordination interactions, π⁃π interactions | Tumor treatment | [ |
| Procyanidin | Fe3+ | Hydrophobic interactions, Coordination interactions, Hydrogen Bond | Colitis | [ |
Table 1 A review of the types of drugs loaded in polyphenol-based supramolecular nanodrug delivery systems, the types of interaction forces involved, and their biomedical applications
| Types of polyphenols | Loaded substance | Supramolecular interaction | Application | Ref. |
|---|---|---|---|---|
| Catechol | DOX, Zn2+ | Hydrophobic interactions, Coordination interactions | Tumor treatment | [ |
| EGCG, Catechol | W6+ | Hydrophobic interactions, Coordination interactions | Tumor treatment | [ |
| Gallic acid, Catechol | Hf4+, Hemoglobin | Hydrophobic interactions, Coordination interactions | Tumor treatment | [ |
| Rosmarinic acid | Dexamethasone | Hydrophobic interactions | Colitis | [ |
| Quercetin | None | Hydrophobic interactions, Hydrogen Bond | Colitis | [ |
| TA | Green fluorescent protein | Hydrophobic interactions | Heart disease | [ |
| Salvianolic acid B, Catechol | Ca2+ | Hydrophobic interactions, Coordination interactions | Renal fibrosis | [ |
| EGCG | Fe3+, DOX | Hydrophobic interactions, Coordination interactions, π⁃π interactions | Tumor treatment | [ |
| Polydopamine | Zn2+, DOX | Coordination interactions | Tumor treatment | [ |
| TA | BSA | Hydrophobic interactions, Hydrogen Bond | Colitis | [ |
| Catechol | 5⁃Aza, W6+ | Hydrophobic interactions, Coordination interactions | Tumor treatment | [ |
| TA | DOX | Hydrophobic interactions, Hydrogen Bond | Tumor treatment | [ |
| EGCG | DOX, Fe3+ | Hydrophobic interactions, Coordination interactions, π⁃π interactions | Tumor treatment | [ |
| Procyanidin | Fe3+ | Hydrophobic interactions, Coordination interactions, Hydrogen Bond | Colitis | [ |
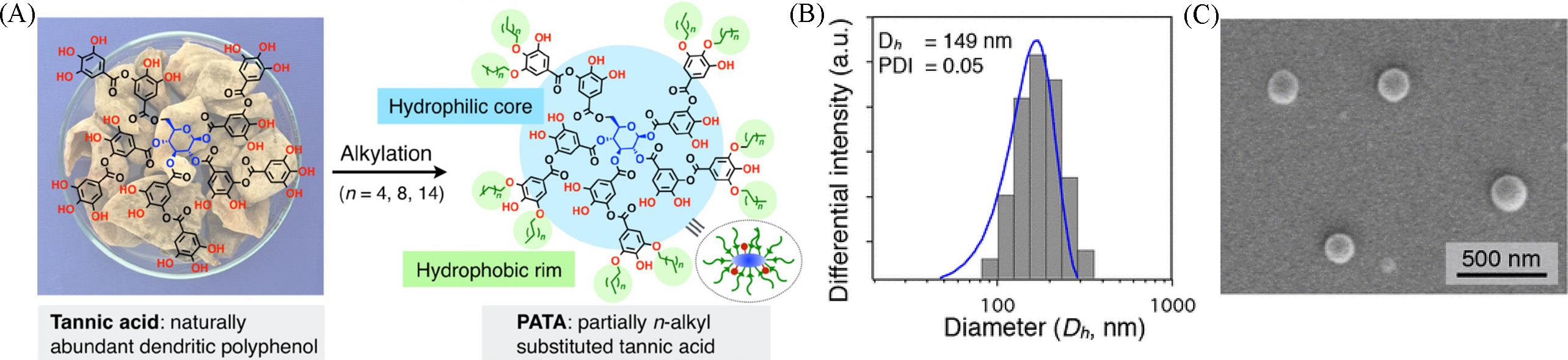
Fig.16 Schematic diagram(A), DLS data(B) and TEM image(C) of the nanoparticles prepared by alkylation of tannic acid[77]Copyright 2018, American Chemical Society.
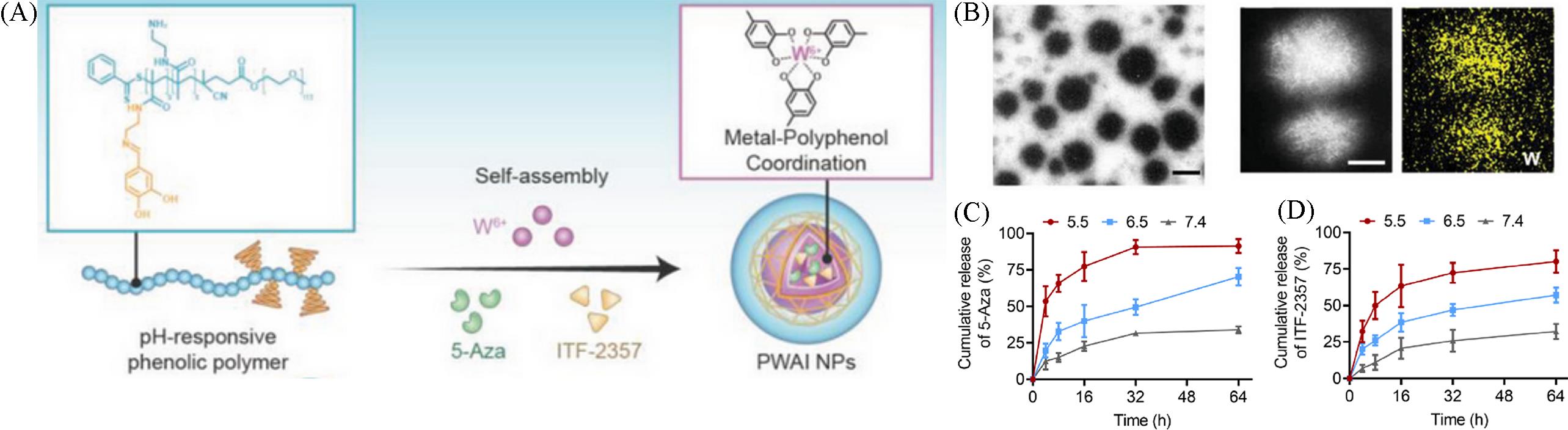
Fig.17 The self⁃assembling(A), TEM image(B) and drug release under different conditions(C, D) of polyphenol⁃containing amphiphilic polymer nanoparticles[17](B) Scale bar: 100 nm; (C, D) red line: pH=5.5; blue line: pH=6.5; grey line: pH=7.4.Copyright 2024, John Wiley and Sons.
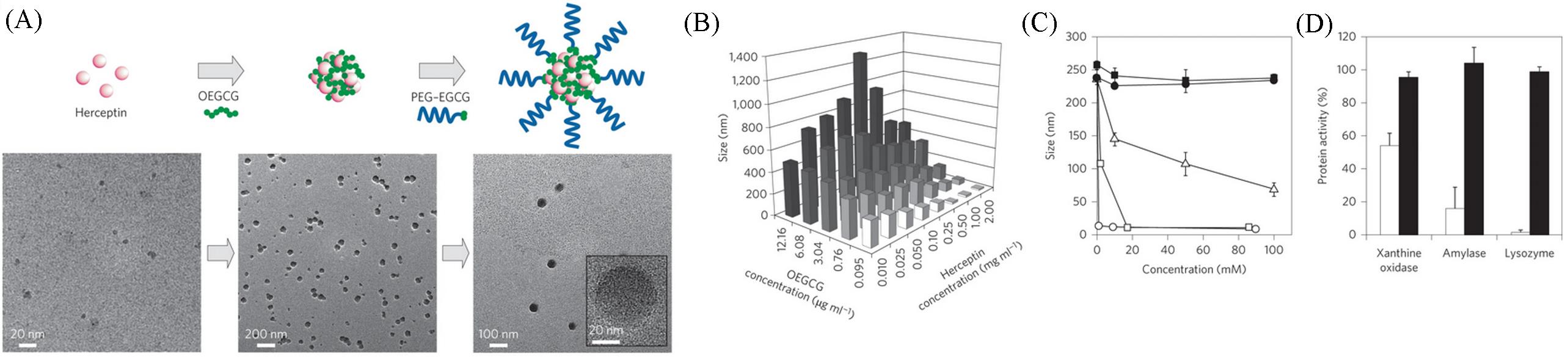
Fig.18 The formation(A), size affected by the concentrations of EGCG and Herceptin(B), dissociation after co⁃incubating with Tween 20(open circles), Triton X⁃100(open squares), SDS(open triangles), urea(filled circles), or NaCl(filled squares)(C) and protein activity inhibition(open bars) and full restoration(solid bars)[m(protein)/m(EGCG)=1∶1](D) of protein(i.e. herceptin)⁃containing EGCG nanoparticles[49]Copyright 2014, Springer Nature.
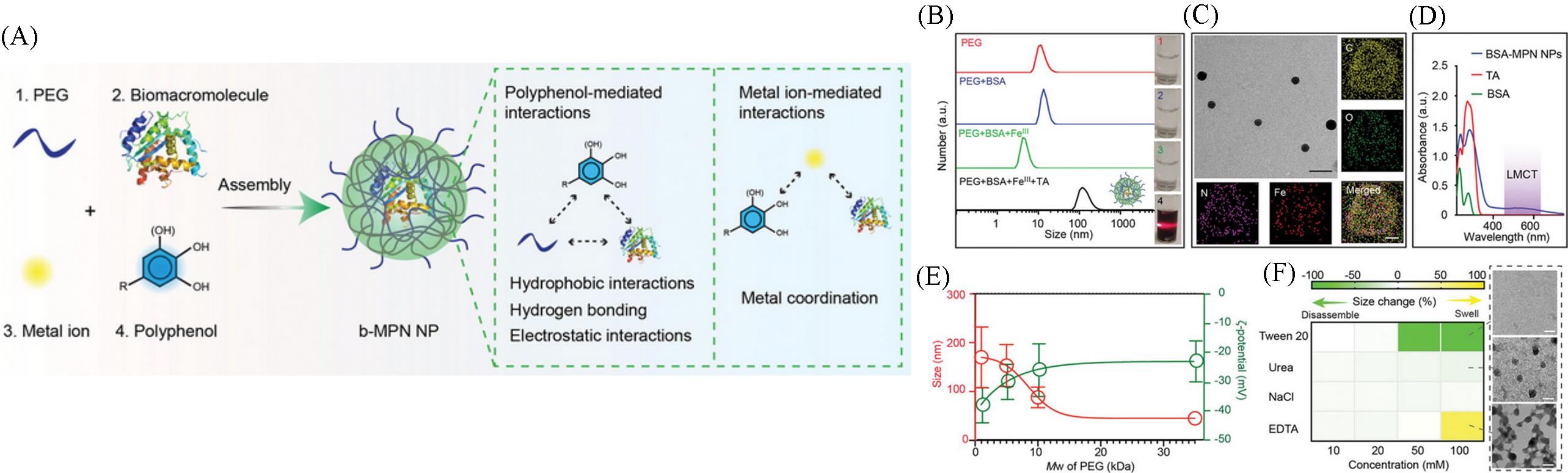
Fig.19 The synthesis(A), size variation(B), morphology(C), characteristic UV⁃Vis spectra(D) of protein⁃containing metal⁃polyphenol⁃protein nanoparticles(b⁃MPN) and their size and zeta potential change when using PEG with different molecular weights(E) and after different solvent incubations(F)[50]Copyright 2022, John Wiley and Sons.
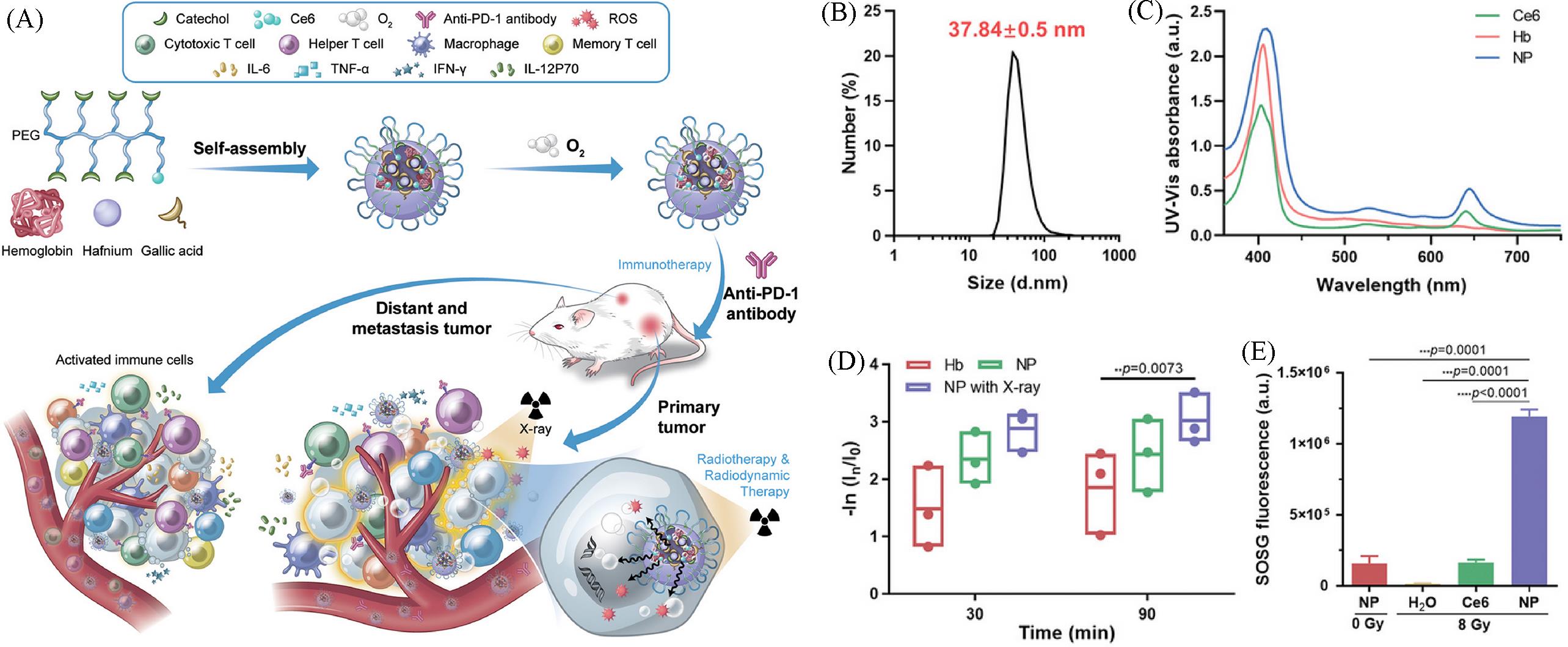
Fig.20 Schematic diagram of the preparation and anti⁃tumor effect(A), particle size distribution(B), UV⁃Vis spectra showing the successful Ce6 loading(C), oxygen carrying capacity(D) and ROS production(E) of hemoglobin⁃loaded polyphenol nanoparticles(Hb@Hf⁃Ce6)[70]Copyright 2021, John Wiley and Sons.
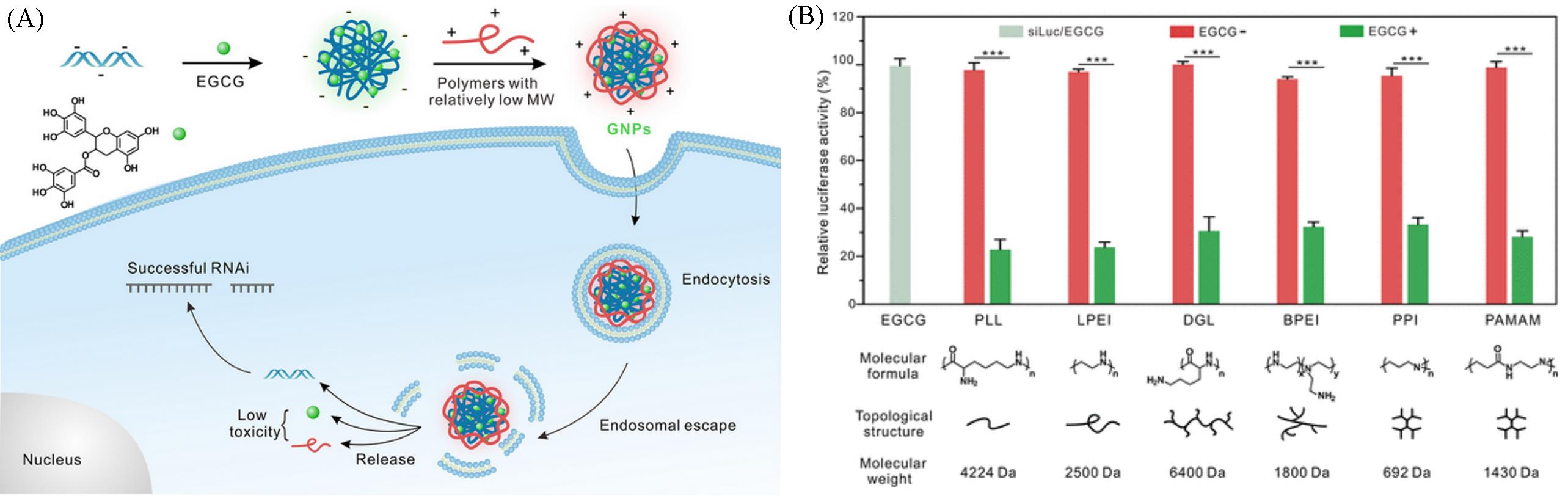
Fig.21 Schematic illustration of the self⁃assembly and gene silencing mechanism of siRNA⁃loaded GNP nanoparticles(A) and the RNAi efficiency of GNPs(green bars) on HeLa⁃Luc cells for 24 h(B)[84]Copyright 2018, American Chemical Society.
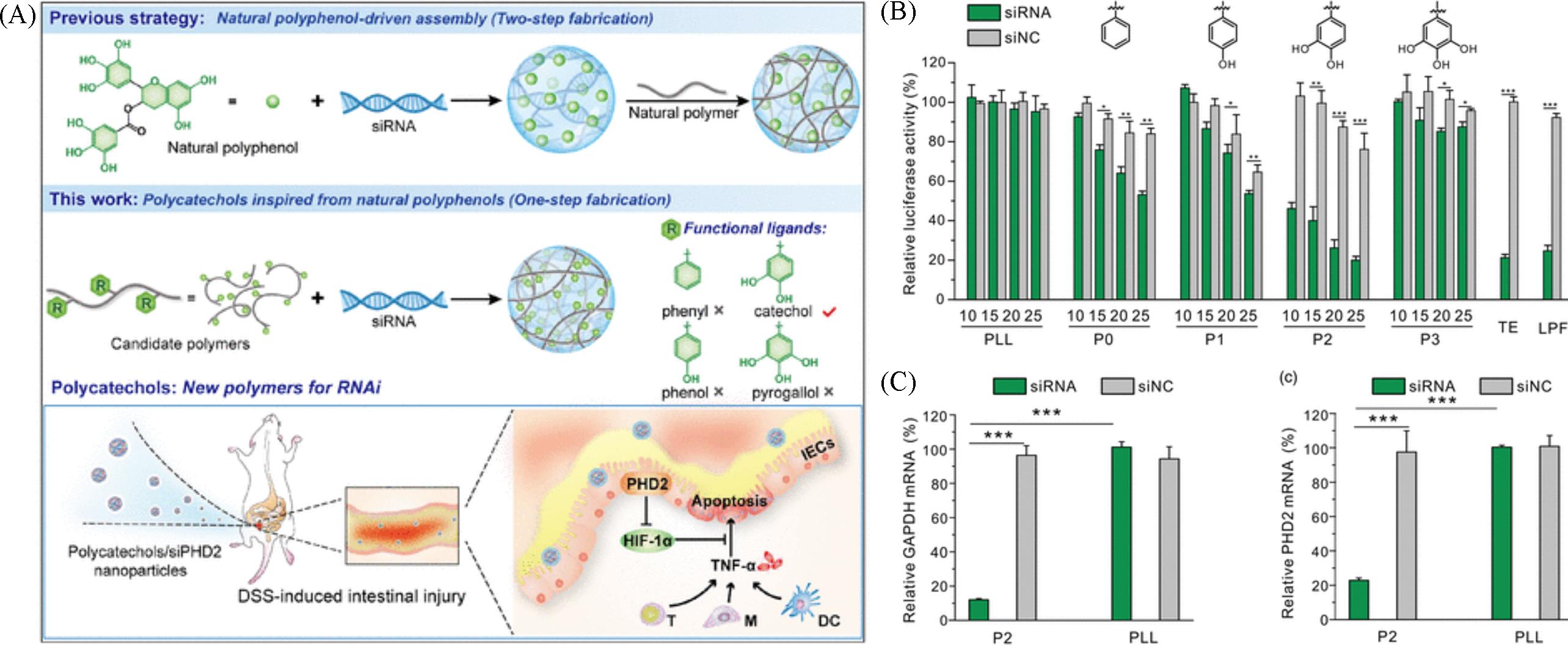
Fig.22 Construction(A), luciferase activity(B), GAPDH(left) and PHD2(right) silencing efficiencies in HeLa cells(C) of siRNA loading polycatechol nanoparticles[85]Copyright 2020, Chinese Chemical Society.
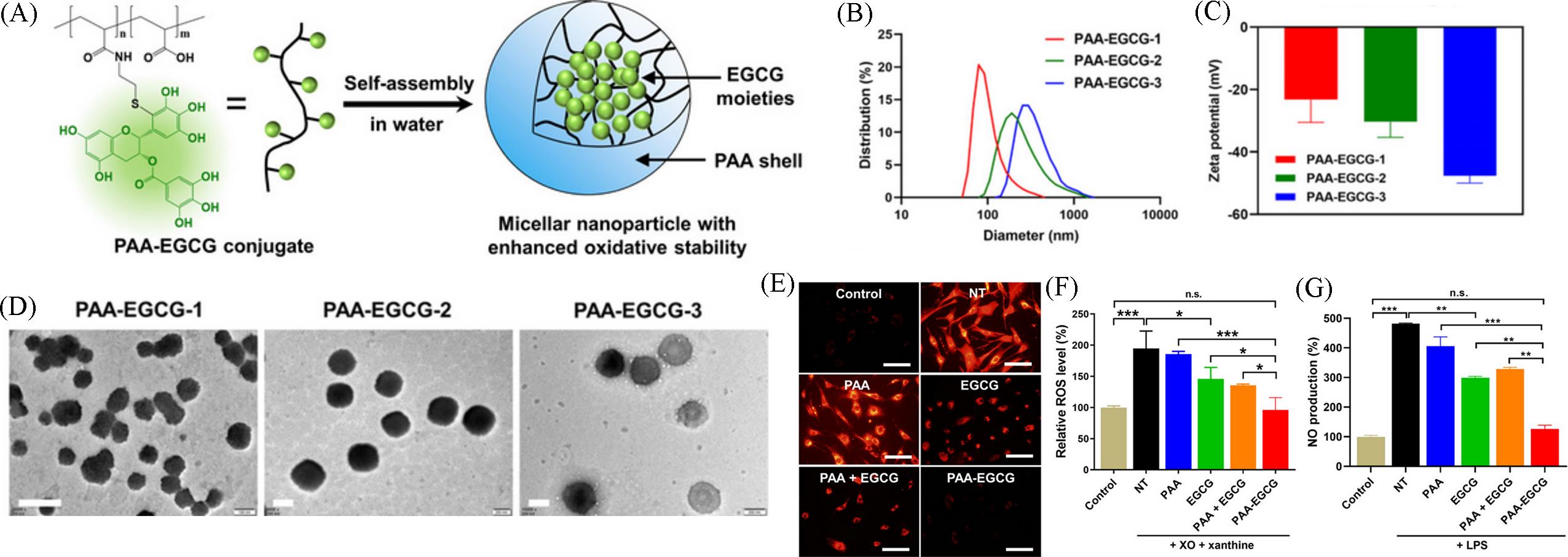
Fig.23 Structure(A), size distribution(B), zeta potential(C), TEM images(D), ROS Brite staining images(E), relative ROS levels(F) and NO production(G) of PAA⁃EGCG nanoparticles with different grafting rates[86]Molar ratio of EGCG to the carboxylic acid group of PAA⁃EGCG: 1.6(PAA⁃EGCG⁃1); 1.0(PAA⁃EGCG⁃2); 0.6(PAA⁃EGCG⁃3).Copyright 2022, American Chemical Society.
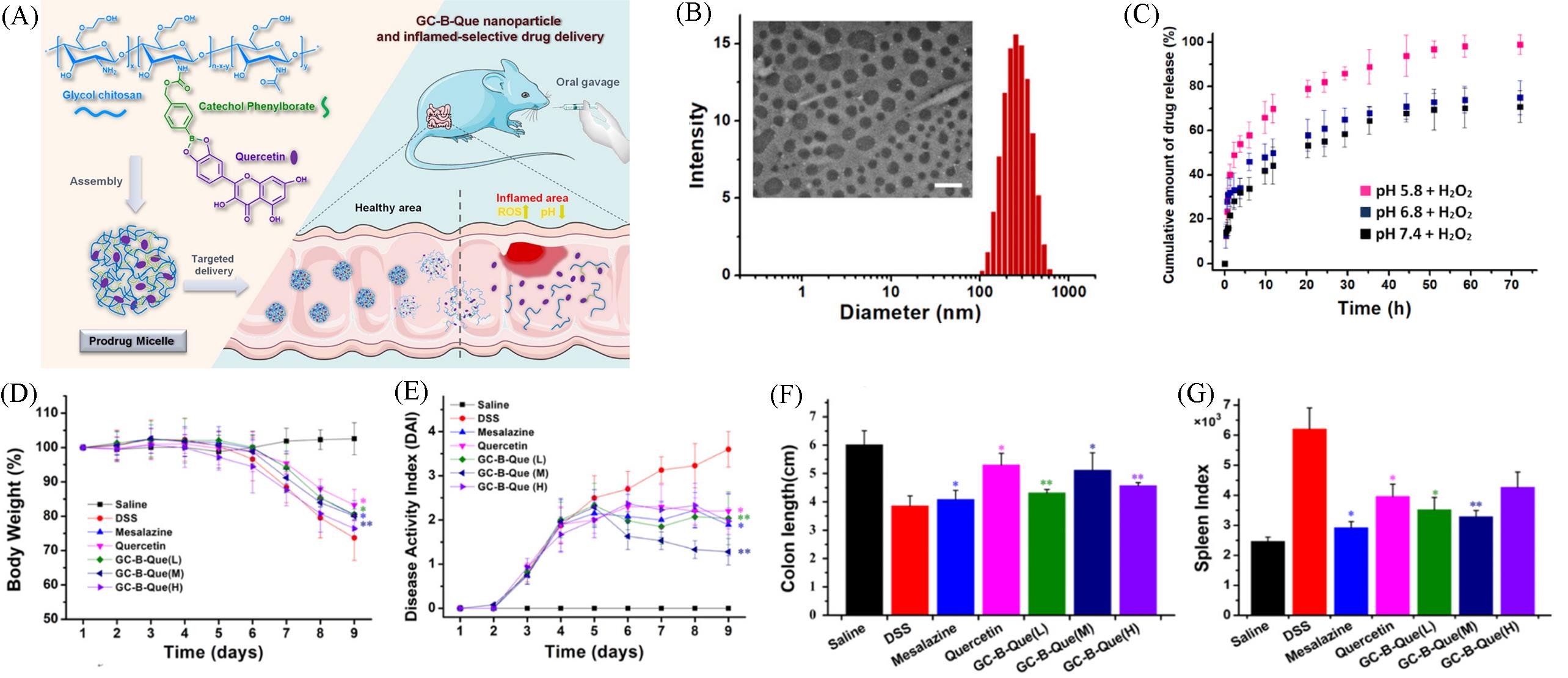
Fig.24 Schematic of the synthesis(A), size distribution and TEM image(B), Que release under different conditions(C) of anti⁃inflammatory GC⁃B⁃Que particles and their treatment effects on mouse body weight(D), disease activity index(E), colon length(F), and spleen index(G) in DSS⁃induced mouse colitis model[66]Copyright 2021, American Chemical Society.
| 1 | De Greef T. F., Meijer E. W., Nature, 2008, 453, 171—173 |
| 2 | De Greef T. F., Smulders M. M., Wolffs M., Schenning A. P., Sijbesma R. P., Meijer E. W., Chem. Rev., 2009, 109, 5687—5754 |
| 3 | Aida T., Meijer E. W., Stupp S. I., Science, 2012, 335, 813—817 |
| 4 | Lehn J. M., Europ. Rev., 2009, 17, 263—280 |
| 5 | Webber M. J., Appel E. A., Meijer E. W., Langer R., Nat. Mater., 2016, 15, 13—26 |
| 6 | Webber M. J., Langer R., Chem. Soc. Rev., 2017, 46, 6600—6620 |
| 7 | Qin B., Yin Z., Tang X., Zhang S., Wu Y., Xu J. F., Zhang X., Prog. Polym. Sci., 2020, 100, 101167 |
| 8 | Peng H. Q., Zhu W., Guo W. J., Li Q., Ma S., Bucher C., Liu B., Ji X., Huang F., Sessler J. L., Prog. Polym. Sci., 2023, 137, 101635 |
| 9 | Liu T., Pan P., Shi H., Feng J., Zhang X. Z., J. Polym. Sci., 2023, 62, 297—323 |
| 10 | Jiang X., Zhang J., Lo P. K., Mao Z., Adv. NanoBiomed Res., 2023, 3, 2200168 |
| 11 | Zhou J., Lin Z., Ju Y., Rahim M. A., Richardson J. J., Caruso F., Acc. Chem. Res., 2020, 53, 1269—1278 |
| 12 | Ejima H., Richardson J. J., Liang K., Best J. P., van Koeverden M. P., Such G. K., Cui J., Caruso F., Science, 2013, 341, 154—157 |
| 13 | Zhou J., Lin Z., Penna M., Pan S., Ju Y., Li S., Han Y., Chen J., Lin G., Richardson J. J., Yarovsky I., Caruso F., Nat. Commun., 2020, 11, 4804 |
| 14 | Guo J., Tardy B. L., Christofferson A. J., Dai Y., Richardson J. J., Zhu W., Hu M., Ju Y., Cui J., Dagastine R. R., Yarovsky I., Caruso F., Nat. Nanotechnol., 2016, 11, 1105—1111 |
| 15 | Xu W., Lin Z., Pan S., Chen J., Wang T., Cortez⁃Jugo C., Caruso F., Angew. Chem. Int. Ed., 2023, 62, e202312925 |
| 16 | Yu X., Li B., Yan J., Li W., Tian H., Wang G., Zhou S., Dai Y., Biomaterials, 2024, 307, 122512 |
| 17 | Wang G., Yan J., Tian H., Li B., Yu X., Feng Y., Li W., Zhou S., Dai Y., Adv. Mater., 2024, 36, e2312588 |
| 18 | Tian Y., Tian H., Li B., Feng C., Dai Y., Small, 2024, 20, e2309850 |
| 19 | Feng Y., Wang G., Li W., Yan J., Yu X., Tian H., Li B., Dai Y., Adv. Healthc. Mater., 2024, 13, e2302811 |
| 20 | Xu Z., Liu G., Zheng L., Wu J., Nano Res., 2022, 16, 905—916 |
| 21 | Li J., Li J., Wei J., Zhu X., Qiu S., Zhao H., ACS Appl. Mater. Interfaces, 2021, 13, 10446—10456 |
| 22 | Venkatesan R., Sivaprakash P., Kim I., Eldesoky G. E., Kim S. C., J. Environ. Chem. Eng., 2023, 11, 110194 |
| 23 | Wu X., Wang L., Tang L., Wang L., Cao S., Wu Q., Zhang Z., Li L., J. Funct. Foods, 2018, 46, 312—319 |
| 24 | Luo R., Lin M., Zhang C., Shi J., Zhang S., Chen Q., Hu Y., Zhang M., Zhang J., Gao F., Food Chem., 2020, 330, 127241 |
| 25 | Chung C. H., Jung W., Keum H., Kim T. W., Jon S., ACS Nano, 2020, 14, 6887—6896 |
| 26 | Qi Y., Li J., Nie Q., Gao M., Yang Q., Li Z., Li Q., Han S., Ding J., Li Y., Zhang J., Biomaterials, 2021, 275, 120952 |
| 27 | Tan H., Sun J., Jin D., Song J., Lei M., Antoshin A., Chen X., Yin M., Qu X., Liu C., Biomater. Sci., 2020, 8, 3334—3347 |
| 28 | Chen R., Zhu C., Xu L., Gu Y., Ren S., Bai H., Zhou Q., Liu X., Lu S., Bi X., Li W., Jia X., Chen Z., Biomaterials, 2021, 274, 120855 |
| 29 | Shin M., Lee H. A., Lee M., Shin Y., Song J. J., Kang S. W., Nam D. H., Jeon E. J., Cho M., Do M., Park S., Lee M. S., Jang J. H., Cho S. W., Kim K. S., Lee H., Nat. Biomed. Eng., 2018, 2, 304—317 |
| 30 | Rahim M. A., Kristufek S. L., Pan S., Richardson J. J., Caruso F., Angew. Chem. Int. Ed. Engl., 2019, 58, 1904—1927 |
| 31 | Guo J., Ping Y., Ejima H., Alt K., Meissner M., Richardson J. J., Yan Y., Peter K., von Elverfeldt D., Hagemeyer C. E., Caruso F., Angew. Chem. Int. Ed., 2014, 53, 5546—5551 |
| 32 | Li S. C., Wang J. G., Jacobson P., Gong X. Q., Selloni A., Diebold U., J. Am. Chem. Soc., 2009, 131, 980—984 |
| 33 | Wei W., Petrone L., Tan Y., Cai H., Israelachvili J. N., Miserez A., Waite J. H., Adv. Funct. Mater., 2016, 26, 3496—3507 |
| 34 | Jankovic I. A., Saponjic Z. V., Dzunuzovic E. S., Nedeljkovic J. M., Nanoscale Res. Lett., 2009, 5, 81—88 |
| 35 | Tian Y., Sang W., Tian H., Xie L., Wang G., Zhang Z., Li W., Dai Y., Adv. Funct. Mater., 2022, 32, 2205690 |
| 36 | Rahim M. A., Ejima H., Cho K. L., Kempe K., Müllner M., Best J. P., Caruso F., Chem. Mater., 2014, 26, 1645—1653 |
| 37 | Muzolf M., Szymusiak H., Gliszczynska⁃Swiglo A., Rietjens I. M., Tyrakowska B., J. Agric. Food Chem., 2008, 56, 816—823 |
| 38 | Kim B. S., Lee H. I., Min Y., Poon Z., Hammond P. T., Chem. Commun. (Camb.), 2009, (28), 4194—4196 |
| 39 | Chen J., Kozlovskaya V., Goins A., Campos⁃Gomez J., Saeed M., Kharlampieva E., Biomacromolecules, 2013, 14, 3830—3841 |
| 40 | Shutava T., Prouty M., Kommireddy D., Lvov Y., Macromolecules, 2005, 38, 2850—2858 |
| 41 | Dierendonck M., Fierens K., de Rycke R., Lybaert L., Maji S., Zhang Z., Zhang Q., Hoogenboom R., Lambrecht B. N., Grooten J., Remon J. P., de Koker S., de Geest B. G., Adv. Funct. Mater., 2014, 24, 4634—4644 |
| 42 | Jakobek L., Food Chem., 2015, 175, 556—567 |
| 43 | Shin M., Ryu J. H., Park J. P., Kim K., Yang J. W., Lee H., Adv. Funct. Mater., 2015, 25, 1270—1278 |
| 44 | Yu J., Wei W., Danner E., Israelachvili J. N., Waite J. H., Adv. Mater., 2011, 23, 2362—2366 |
| 45 | Fan Q., Yang Z., Li Y., Cheng Y., Li Y., Adv. Funct. Mater., 2021, 31, 2101646 |
| 46 | Quan T. H., Benjakul S., Sae⁃leaw T., Balange A. K., Maqsood S., Trends in Food Sci. & Tech., 2019, 91, 507—517 |
| 47 | Dai S., Xu T., Yuan Y., Fang Q., Lian Z., Tian T., Tong X., Jiang L., Wang H., Food Hydrocolloids, 2024, 146, 109197 |
| 48 | Charlton A. J., Baxter N. J., Khan M. L., Moir A. J., Haslam E., Davies A. P., Williamson M. P., J. Agric. Food Chem., 2002, 50, 1593—1601 |
| 49 | Chung J. E., Tan S., Gao S. J., Yongvongsoontorn N., Kim S. H., Lee J. H., Choi H. S., Yano H., Zhuo L., Kurisawa M., Ying J. Y., Nat. Nanotechnol., 2014, 9, 907—912 |
| 50 | Chen J., Pan S., Zhou J., Lin Z., Qu Y., Glab A., Han Y., Richardson J. J., Caruso F., Adv. Mater., 2022, 34, e2108624 |
| 51 | Zhang C., Hu D. F., Xu J. W., Ma M. Q., Xing H., Yao K., Ji J., Xu Z. K., ACS Nano, 2018, 12, 12347—12356 |
| 52 | Tian H., Wang G., Sang W., Xie L., Zhang Z., Li W., Yan J., Tian Y., Li J., Li B., Dai Y., Nano Today, 2022, 43, 101405 |
| 53 | Lv M., Chen M., Zhang R., Zhang W., Wang C., Zhang Y., Wei X., Guan Y., Liu J., Feng K., Jing M., Wang X., Liu Y. C., Mei Q., Han W., Jiang Z., Cell Res., 2020, 30, 966—979 |
| 54 | Mandal M. K., Domb A. J., Pharmaceutics, 2024, 16, 718 |
| 55 | Fang R., Hao R., Wu X., Li Q., Leng X., Jing H., J. Agric. Food Chem., 2011, 59, 6292—6298 |
| 56 | Ren P. F., Yang H. C., Liang H. Q., Xu X. L., Wan L. S., Xu Z. K., Langmuir, 2015, 31, 5851—5858 |
| 57 | Liu S., Ji W., Wu T., He Y., Huang Y., Yu Y., Yu W., ACS Sustainable Chem. Eng., 2024, 12, 4224—4235 |
| 58 | Gao Z., Zharov I., Chem. Mater., 2014, 26, 2030—2037 |
| 59 | Li W., Yan J., Tian H., Li B., Wang G., Sang W., Zhang Z., Zhang X., Dai Y., Bioact. Mater., 2023, 22, 34—46 |
| 60 | Martinez C. R., Iverson B. L., Chem. Sci., 2012, 3, 2191—2201 |
| 61 | Liu F. F., Fan J. L., Wang S. G., Ma G. H., Chem. Eng. J., 2013, 219, 450—458 |
| 62 | Lin D., Xing B., Environ. Sci. Technol., 2008, 42, 5917—5923 |
| 63 | Luo J., Lai J., Zhang N., Liu Y., Liu R., Liu X., ACS Sustainable Chem. Eng., 2016, 4, 1404—1413 |
| 64 | Liang K., Chung J. E., Gao S. J., Yongvongsoontorn N., Kurisawa M., Adv. Mater., 2018, 30, e1706963 |
| 65 | Philp D., Stoddart J. F., Angew. Chem. Int. Ed., 1996, 35(11), 1154—1196 |
| 66 | Shen C., Zhao L., Du X., Tian J., Yuan Y., Jia M., He Y., Zeng R., Qiao R., Li C., Mol. Pharm., 2021, 18, 1419—1430 |
| 67 | Liu Z., Yu W., Sheng W., Li R., Guo H., Feng X., Li Q., Wang R., Li W., Jia X., Biomacromolecules, 2022, 23, 140—149 |
| 68 | Xu C., Wang Y., Yu H., Tian H., Chen X., ACS Nano, 2018, 12, 8255—8265 |
| 69 | Wang G., Li B., Tian H., Xie L., Yan J., Sang W., Li J., Zhang Z., Li W., Dai Y., Adv. Funct. Mater., 2023, 33, 2213425 |
| 70 | Sang W., Xie L., Wang G., Li J., Zhang Z., Li B., Guo S., Deng C. X., Dai Y., Adv. Sci.(Weinh), 2021, 8, 2003338 |
| 71 | Li J., Zhang C., He W., Qiao H., Chen J., Wang K., Oupicky D., Sun M., Biomater. Sci., 2017, 6, 179—188 |
| 72 | Shi H., Wang R., Cao H. C., Guo H. Y., Pan P., Xiong C. F., Zhang L. J., Yang Q., Wei S., Liu T., Adv. Healthc. Mater., 2023, 12, e2300054 |
| 73 | Zhan L., Yin X., Zhang Y., Ju J., Wu Y., Ding L., Li C., Chen X., Wang Y., Biomater. Adv., 2023, 146, 213306 |
| 74 | Wang X., Yan J., Wang L., Pan D., Xu Y., Wang F., Sheng J., Li X., Yang M., Theranostics, 2020, 10, 10808—10822 |
| 75 | Shan L., Gao G., Wang W., Tang W., Wang Z., Yang Z., Fan W., Zhu G., Zhai K., Jacobson O., Dai Y., Chen X., Biomaterials, 2019, 210, 62—69 |
| 76 | Zhao Y., Xu L., Kong F., Yu L., Chem. Eng. J., 2021, 416, 129090 |
| 77 | Payra D., Yamauchi Y., Samitsu S., Naito M., Chem. Mater., 2018, 30, 8025—8033 |
| 78 | Nagesh P. K. B., Chowdhury P., Hatami E., Kumari S., Kashyap V. K., Tripathi M. K., Wagh S., Meibohm B., Chauhan S. C., Jaggi M., Yallapu M. M., ACS Appl. Mater. Interf., 2019, 11, 38537—38554 |
| 79 | Patel A. R., Seijen⁃ten⁃Hoorn J., Heussen P. C. M., Drost E., Hazekamp J., Velikov K. P., J. Colloid Interf. Sci., 2012, 374, 150—156 |
| 80 | Von Staszewski M., Jara F. L., Ruiz A. L. T. G., Jagus R. J., Carvalho J. E., Pilosof A. M. R., J. Funct. Foods, 2012, 4, 800—809 |
| 81 | Zheng X., Chen A., Hoshi T., Anzai J., Li G., Anal. Bioanal. Chem., 2006, 386, 1913—1919 |
| 82 | Kuzuhara T., Sei Y., Yamaguchi K., Suganuma M., Fujiki H., J. Biol. Chem., 2006, 281, 17446—17456 |
| 83 | Galindo⁃Murillo R., Cheatham III T. E., J. Biomol. Struct. Dyn., 2018, 36, 3311—3323 |
| 84 | Shen W., Wang Q., Shen Y., Gao X., Li L., Yan Y., Wang H., Cheng Y., ACS Cent. Sci., 2018, 4, 1326—1333 |
| 85 | Shen W., Wang R., Fan Q., Gao X., Wang H., Shen Y., Li Y., Cheng Y., CCS Chemistry, 2020, 2, 146—157 |
| 86 | Bae K. H., Chan K. H., Kurisawa M., ACS Macro. Lett., 2022, 11, 835—840 |
| [1] | 梁惠闲, 王淼, 张雨璨, 白丽, 李雪妹, 于法标, 程子译, 赵琳璐. 基于Ag2S量子点的光热治疗协同药物治疗在动脉粥样硬化中的应用[J]. 高等学校化学学报, 2025, 46(1): 196. |
| [2] | 萨仁格日乐, 梁靖, 关少钰, 常宇佳, 张佳博, 梁楠, 亢静, 董阿力德尔图. N-卤胺聚合物表面修饰金纳米粒子的抗菌性能[J]. 高等学校化学学报, 2025, 46(1): 252. |
| [3] | 宁佳雨, 郝鹏飞, 王峰, 叶家全, 崇羽. 功能性多酚-精氨酸自组装纳米药物用于乳腺癌放射增敏[J]. 高等学校化学学报, 2025, 46(1): 206. |
| [4] | 陈俊年, 张海峰, 王海兵, 杨火诚, 罗忠. 基于超分子纳米递送系统的精准医疗研究进展[J]. 高等学校化学学报, 2025, 46(1): 50. |
| [5] | 张荡, 孙小敏, 杨海跃, 宋勃翰, 丛萌, 王宇新, 丁锋, 徐珊珊, 毕赛, 王磊. 基于纳米酶的微纳米马达在智能药物递送中的应用[J]. 高等学校化学学报, 2025, 46(1): 76. |
| [6] | 乐鑫, 贾庭芳, 陈瑶, 周远柱, 曲佳菲, 李聪, 申静. 稀土上转换纳米粒子掺杂口腔复合树脂材料的近红外光固化[J]. 高等学校化学学报, 2025, 46(1): 188. |
| [7] | 李滨汐, 张燕, 姚栋. 具有磁共振/荧光双模式成像功能的Fe3O4/CuInS2二元超粒子[J]. 高等学校化学学报, 2025, 46(1): 234. |
| [8] | 孙玉洁, 牛振田, 佟昊轩, 胡悦, 俞丙然, 徐福建. 开环制备聚二硫化物及其在药物递送方面的应用[J]. 高等学校化学学报, 2025, 46(1): 20240116. |
| [9] | 曾自强, 张晨杰, 徐敏敏, 姚建林. Au纳米粒子二聚体-Au片耦合体系“热点”的尺寸效应[J]. 高等学校化学学报, 2024, 45(6): 20240022. |
| [10] | 马勤政, 王伟, 梁旭婷. 石墨烯-金纳米材料修饰电极用于L-酪氨酸的检测[J]. 高等学校化学学报, 2024, 45(4): 20230521. |
| [11] | 闫勇杰, 高文博, 鲁晨辉, 杨成, 徐姝婷. 基于微萃取-纳喷雾质谱技术的纳升脑脊液中咖啡多酚的检测[J]. 高等学校化学学报, 2024, 45(11): 20240327. |
| [12] | 胡文馨, 赵莹, 杜丹阳, 张红丹, 程鹏. ZSM-5封装Pt-La双金属催化剂的制备及对异丁烷裂解反应的催化性能[J]. 高等学校化学学报, 2024, 45(10): 20240244. |
| [13] | 朱润芝, 王怡, 纳佳雪, 曹乐乐, 张辉, 王迎辉, 孟哲. 基于光诱导电子转移的比率型发光传感器定量检测生物样本中的多巴胺[J]. 高等学校化学学报, 2024, 45(1): 20230391. |
| [14] | 闫雨甜, 吴思, 常康康, 夏宇正, 陈晓农, 石淑先. 表面覆盖型AuNPs@PNIPAM复合粒子的制备及催化性能[J]. 高等学校化学学报, 2023, 44(4): 20220636. |
| [15] | 盛劲菡, 郑琪臻, 汪铭. CRISPR/Cas9基因编辑非病毒递送系统[J]. 高等学校化学学报, 2023, 44(3): 20220344. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||