

高等学校化学学报 ›› 2018, Vol. 39 ›› Issue (7): 1540.doi: 10.7503/cjcu20170657
刘海春1, 卢帅1, 张艳敏1, 周伟能1, 尹凌枫1, 朱露1, 赵珺楠1, 陆涛1,2( ), 陈亚东1(
), 陈亚东1( )
)
收稿日期:2017-09-29
出版日期:2018-07-10
发布日期:2018-06-19
作者简介:联系人简介: 陆 涛, 男, 博士, 教授, 主要从事新药开发研究. E-mail:基金资助:
LIU Haichun1, LU Shuai1, ZHANG Yanmin1, ZHOU Weineng1, YIN Lingfeng1, ZHU Lu1, ZHAO Junnan1, LU Tao1,2,*( ), CHEN Yadong1,*(
), CHEN Yadong1,*( )
)
Received:2017-09-29
Online:2018-07-10
Published:2018-06-19
Contact:
LU Tao,CHEN Yadong
E-mail:lutao@cpu.edu.cn;ydchen@cpu.edu.cn
Supported by:摘要:
通过动力学模拟获得JAK2高选择性抑制剂Fedratinib在JAK2和JAK3激酶中的结合构象, 结合自由能的计算结果表明Fedratinib在JAK2中更稳定. 将能量分解到结合位点氨基酸, 分析发现当分子在JAK2中占据P-loop区的疏水口袋, 并与附近Arg980和Asp994等氨基酸形成氢键时, 可以增加相对于JAK2的选择性.
中图分类号:
TrendMD:
刘海春, 卢帅, 张艳敏, 周伟能, 尹凌枫, 朱露, 赵珺楠, 陆涛, 陈亚东. 分子动力学模拟研究Fedratinib-JAK2/JAK3选择性. 高等学校化学学报, 2018, 39(7): 1540.
LIU Haichun, LU Shuai, ZHANG Yanmin, ZHOU Weineng, YIN Lingfeng, ZHU Lu, ZHAO Junnan, LU Tao, CHEN Yadong. Molecular Dynamics Simulation of the Selectivity of Fedratinib Complex with JAK2/JAK3†. Chem. J. Chinese Universities, 2018, 39(7): 1540.
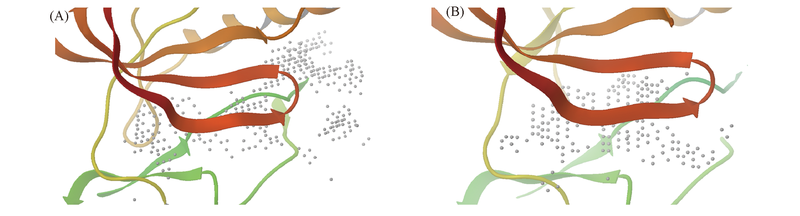
Fig.1 Comparison of “site points” for the ligand binding site in X-ray structures of JAK2(A) and JAK3(B) generated by SiteMap(depicted with white spheres)
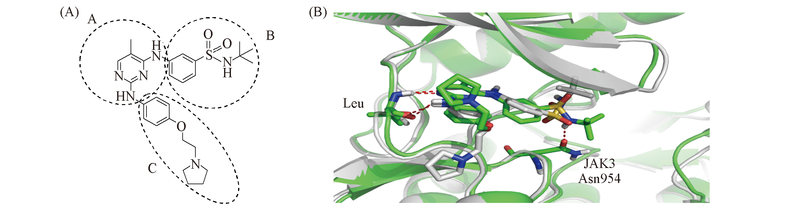
Fig.2 Docking interaction diagram of Fedratinib into the binding site of JAK2(white) and JAK3(green)^ (A) Structure of Fedratinib; (B) binding modes. The hydrogen bonding interactions were depicted with red dotted lines.
| Enzyme | Acceptor | Donor | Distance/nm | Angle/(°) | Occupied(%) |
|---|---|---|---|---|---|
| JAK2 | Fed@O3 | 980@NH2: HH21 | 0.292(±0.014) | 18.58(±9.75) | 99.34 |
| Fed@N2 | 932@N: H | 0.311(±0.014) | 18.14(±9.65) | 98.34 | |
| 932@O | Fed@N3: H2 | 0.302(±0.016) | 30.67(±11.75) | 96.26 | |
| Fed@O2 | 980@NE: HE | 0.311(±0.019) | 35.50(±13.31) | 71.75 | |
| 994@OD1 | Fed@N6: HH27 | 0.305(±0.019) | 21.96(±12.61) | 50.37 | |
| Fed@O2 | 980@NH2: HH21 | 0.329(±0.016) | 47.30(±8.60) | 23.14 | |
| 994@OD2 | Fed@N6: H27 | 0.308(±0.020) | 20.68(±11.45) | 22.30 | |
| Fed@O3 | 980@NE: HE | 0.333(±0.014) | 43.87(±7.05) | 19.98 | |
| JAK3 | Fed@N2 | 905@N: H | 0.314(±0.014) | 17.23(±9.48) | 97.42 |
| 905@O | Fed@N3: H3 | 0.301(±0.016) | 34.29(±12.68) | 92.76 | |
| Fed@O3 | 830@N: H | 0.315(±0.019) | 50.26(±8.20) | 5.42 | |
| Fed@O2 | 855@NZ: HZ1 | 0.328(±0.014) | 43.66(±6.41) | 0.10 |
Table 1 Hydrogen bonds analysis of JAK2/JAK3-Fedratinib from the MD trajectories
| Enzyme | Acceptor | Donor | Distance/nm | Angle/(°) | Occupied(%) |
|---|---|---|---|---|---|
| JAK2 | Fed@O3 | 980@NH2: HH21 | 0.292(±0.014) | 18.58(±9.75) | 99.34 |
| Fed@N2 | 932@N: H | 0.311(±0.014) | 18.14(±9.65) | 98.34 | |
| 932@O | Fed@N3: H2 | 0.302(±0.016) | 30.67(±11.75) | 96.26 | |
| Fed@O2 | 980@NE: HE | 0.311(±0.019) | 35.50(±13.31) | 71.75 | |
| 994@OD1 | Fed@N6: HH27 | 0.305(±0.019) | 21.96(±12.61) | 50.37 | |
| Fed@O2 | 980@NH2: HH21 | 0.329(±0.016) | 47.30(±8.60) | 23.14 | |
| 994@OD2 | Fed@N6: H27 | 0.308(±0.020) | 20.68(±11.45) | 22.30 | |
| Fed@O3 | 980@NE: HE | 0.333(±0.014) | 43.87(±7.05) | 19.98 | |
| JAK3 | Fed@N2 | 905@N: H | 0.314(±0.014) | 17.23(±9.48) | 97.42 |
| 905@O | Fed@N3: H3 | 0.301(±0.016) | 34.29(±12.68) | 92.76 | |
| Fed@O3 | 830@N: H | 0.315(±0.019) | 50.26(±8.20) | 5.42 | |
| Fed@O2 | 855@NZ: HZ1 | 0.328(±0.014) | 43.66(±6.41) | 0.10 |
| Contribution | JAK2 | JAK3 | ||
|---|---|---|---|---|
| Mean/(kJ·mol-1) | Std./(kJ·mol-1) | Mean/(kJ·mol-1) | Std./(kJ·mol-1) | |
| ΔEele/(kJ·mol-1) | -130.79 | 5.14 | -87.99 | 3.23 |
| ΔEvdw/(kJ·mol-1) | -254.09 | 3.26 | -237.19 | 3.35 |
| ΔEintra/(kJ·mol-1) | 0 | 0 | 0 | 0 |
| ΔEgas/(kJ·mol-1) | -384.89 | 5.80 | -325.18 | 4.31 |
| ΔGnp/(kJ·mol-1) | -32.09 | 0.23 | -32.13 | 0.60 |
| ΔGpb/(kJ·mol-1) | 221.67 | 4.35 | 206.35 | 4.21 |
| ΔGpbele/(kJ·mol-1) | 90.88 | 4.57 | 118.41 | 4.22 |
| ΔGpb,tot/(kJ·mol-1) | -195.31 | 5.04 | -150.92 | 4.63 |
| ΔGgb/(kJ·mol-1) | 180.96 | 2.85 | 168.74 | 2.82 |
| ΔGgbele/(kJ·mol-1) | 50.17 | 3.39 | 80.75 | 2.30 |
| ΔGgb,tot/(kJ·mol-1) | -236.02 | 4.23 | -188.53 | 3.26 |
Table 2 Calculated binding free energies of JAK2/JAK3-Fedratinib using MM/GBSA and MM/PBSA method*
| Contribution | JAK2 | JAK3 | ||
|---|---|---|---|---|
| Mean/(kJ·mol-1) | Std./(kJ·mol-1) | Mean/(kJ·mol-1) | Std./(kJ·mol-1) | |
| ΔEele/(kJ·mol-1) | -130.79 | 5.14 | -87.99 | 3.23 |
| ΔEvdw/(kJ·mol-1) | -254.09 | 3.26 | -237.19 | 3.35 |
| ΔEintra/(kJ·mol-1) | 0 | 0 | 0 | 0 |
| ΔEgas/(kJ·mol-1) | -384.89 | 5.80 | -325.18 | 4.31 |
| ΔGnp/(kJ·mol-1) | -32.09 | 0.23 | -32.13 | 0.60 |
| ΔGpb/(kJ·mol-1) | 221.67 | 4.35 | 206.35 | 4.21 |
| ΔGpbele/(kJ·mol-1) | 90.88 | 4.57 | 118.41 | 4.22 |
| ΔGpb,tot/(kJ·mol-1) | -195.31 | 5.04 | -150.92 | 4.63 |
| ΔGgb/(kJ·mol-1) | 180.96 | 2.85 | 168.74 | 2.82 |
| ΔGgbele/(kJ·mol-1) | 50.17 | 3.39 | 80.75 | 2.30 |
| ΔGgb,tot/(kJ·mol-1) | -236.02 | 4.23 | -188.53 | 3.26 |
| Residue | ΔEvdw/(kJ·mol-1) | ΔEele/(kJ·mol-1) | ΔEgas/(kJ·mol-1) | ΔGgb,tot/(kJ·mol-1) | |||||
|---|---|---|---|---|---|---|---|---|---|
| JAK2 | JAK3 | JAK2 | JAK3 | JAK2 | JAK3 | JAK2 | JAK3 | JAK2 | JAK3 |
| Met929 | Met902 | -2.80 | -2.30 | 1.42 | 0.59 | -1.38 | -1.76 | -2.01 | -1.88 |
| Val911 | Val884 | -2.09 | -1.46 | 0.67 | 0.67 | -1.42 | -0.79 | -1.84 | -1.34 |
Table 3 Decomposition of the binding energy on per residue of GateKeeper basis on two systems JAK2-Fedratinib and JAK3-Fedratinib
| Residue | ΔEvdw/(kJ·mol-1) | ΔEele/(kJ·mol-1) | ΔEgas/(kJ·mol-1) | ΔGgb,tot/(kJ·mol-1) | |||||
|---|---|---|---|---|---|---|---|---|---|
| JAK2 | JAK3 | JAK2 | JAK3 | JAK2 | JAK3 | JAK2 | JAK3 | JAK2 | JAK3 |
| Met929 | Met902 | -2.80 | -2.30 | 1.42 | 0.59 | -1.38 | -1.76 | -2.01 | -1.88 |
| Val911 | Val884 | -2.09 | -1.46 | 0.67 | 0.67 | -1.42 | -0.79 | -1.84 | -1.34 |
| Residue | ΔEvdw/(kJ·mol-1) | ΔEele/(kJ·mol-1) | ΔEgas/(kJ·mol-1) | ΔGgb,tot/(kJ·mol-1) | |||||
|---|---|---|---|---|---|---|---|---|---|
| JAK2 | JAK3 | JAK2 | JAK3 | JAK2 | JAK3 | JAK2 | JAK3 | JAK2 | JAK3 |
| Glu930 | Glu903 | -0.71 | -1.38 | -9.29 | -8.79 | -10.00 | -10.17 | -1.67 | -1.42 |
| Leu932 | Leu905 | -5.73 | -5.56 | -12.84 | -11.51 | -18.58 | -17.07 | -11.63 | -10.63 |
Table 4 Decomposition of the binding energy on per residue of hinge region basis on two systems JAK2-Fedratinib and JAK3-Fedratinib
| Residue | ΔEvdw/(kJ·mol-1) | ΔEele/(kJ·mol-1) | ΔEgas/(kJ·mol-1) | ΔGgb,tot/(kJ·mol-1) | |||||
|---|---|---|---|---|---|---|---|---|---|
| JAK2 | JAK3 | JAK2 | JAK3 | JAK2 | JAK3 | JAK2 | JAK3 | JAK2 | JAK3 |
| Glu930 | Glu903 | -0.71 | -1.38 | -9.29 | -8.79 | -10.00 | -10.17 | -1.67 | -1.42 |
| Leu932 | Leu905 | -5.73 | -5.56 | -12.84 | -11.51 | -18.58 | -17.07 | -11.63 | -10.63 |
| Residue | ΔEvdw/(kJ·mol-1) | ΔEele/(kJ·mol-1) | ΔEgas/(kJ·mol-1) | ΔGgb,tot/(kJ·mol-1) | |||||
|---|---|---|---|---|---|---|---|---|---|
| JAK2 | JAK3 | JAK2 | JAK3 | JAK2 | JAK3 | JAK2 | JAK3 | JAK2 | JAK3 |
| Gly856 | Gly829 | -4.23 | -5.10 | -0.79 | -3.05 | -5.02 | -8.20 | -2.85 | -4.73 |
| Lys857 | Lys830 | -3.35 | -5.23 | 3.35 | -10.25 | 0 | -15.44 | -0.21 | -3.31 |
| Gly858 | Gly831 | -1.55 | -3.89 | 0.29 | 4.27 | -1.26 | 0.38 | -0.29 | 0.13 |
| Gly861 | Gly834 | -2.55 | -0.67 | 0.46 | 0.88 | -2.05 | 0.21 | -1.26 | -0.67 |
| Ser862 | Ser835 | -2.59 | -0.75 | -0.54 | -1.21 | -3.14 | -1.97 | -2.59 | -0.96 |
| Val863 | Val836 | -10.38 | -7.32 | 0.04 | 1.46 | -10.33 | -5.82 | -11.88 | -7.91 |
Table 5 Decomposition of the binding energy on per residue of P-loop basis on two systems JAK2-Fedratinib and JAK3-Fedratinib
| Residue | ΔEvdw/(kJ·mol-1) | ΔEele/(kJ·mol-1) | ΔEgas/(kJ·mol-1) | ΔGgb,tot/(kJ·mol-1) | |||||
|---|---|---|---|---|---|---|---|---|---|
| JAK2 | JAK3 | JAK2 | JAK3 | JAK2 | JAK3 | JAK2 | JAK3 | JAK2 | JAK3 |
| Gly856 | Gly829 | -4.23 | -5.10 | -0.79 | -3.05 | -5.02 | -8.20 | -2.85 | -4.73 |
| Lys857 | Lys830 | -3.35 | -5.23 | 3.35 | -10.25 | 0 | -15.44 | -0.21 | -3.31 |
| Gly858 | Gly831 | -1.55 | -3.89 | 0.29 | 4.27 | -1.26 | 0.38 | -0.29 | 0.13 |
| Gly861 | Gly834 | -2.55 | -0.67 | 0.46 | 0.88 | -2.05 | 0.21 | -1.26 | -0.67 |
| Ser862 | Ser835 | -2.59 | -0.75 | -0.54 | -1.21 | -3.14 | -1.97 | -2.59 | -0.96 |
| Val863 | Val836 | -10.38 | -7.32 | 0.04 | 1.46 | -10.33 | -5.82 | -11.88 | -7.91 |
| Residue | ΔEvdw/(kJ·mol-1) | ΔEele/(kJ·mol-1) | ΔEgas/(kJ·mol-1) | ΔGgb,tot/(kJ·mol-1) | |||||
|---|---|---|---|---|---|---|---|---|---|
| JAK2 | JAK3 | JAK2 | JAK3 | JAK2 | JAK3 | JAK2 | JAK3 | JAK2 | JAK3 |
| Lys882 | Lys855 | -3.51 | -1.26 | 12.93 | -6.95 | 9.41 | -8.20 | 3.68 | 1.42 |
| Arg980 | Arg953 | -6.99 | -4.39 | -36.94 | 2.09 | -43.93 | -2.26 | -10.04 | 1.34 |
| Asp994 | Asp967 | -6.82 | -4.39 | -17.20 | 2.64 | -24.02 | -1.76 | -4.98 | -2.38 |
Table 6 Decomposition of the binding energy on some other residue basis on two systems JAK2-Fedratinib and JAK3-Fedratinib
| Residue | ΔEvdw/(kJ·mol-1) | ΔEele/(kJ·mol-1) | ΔEgas/(kJ·mol-1) | ΔGgb,tot/(kJ·mol-1) | |||||
|---|---|---|---|---|---|---|---|---|---|
| JAK2 | JAK3 | JAK2 | JAK3 | JAK2 | JAK3 | JAK2 | JAK3 | JAK2 | JAK3 |
| Lys882 | Lys855 | -3.51 | -1.26 | 12.93 | -6.95 | 9.41 | -8.20 | 3.68 | 1.42 |
| Arg980 | Arg953 | -6.99 | -4.39 | -36.94 | 2.09 | -43.93 | -2.26 | -10.04 | 1.34 |
| Asp994 | Asp967 | -6.82 | -4.39 | -17.20 | 2.64 | -24.02 | -1.76 | -4.98 | -2.38 |
| [1] | Takemoto S., Mulloy J. C., Cereseto A., Migone T. S., Patel B. K. R., Matsuoka M., Yamaguchi K., Takatsuki K., Kamihira S., White J. D., Leonard W. J., Waldmann T., Franchini G., Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 13897—13902 |
| [2] | Ivashkiv L. B., Hu X., Arthritis Rheum., 2003, 48(8), 2092—2096 |
| [3] | Yamaoka K., Saharinen P., Pesu M., Holt V. E. Ⅲ, Silvennoinen O., O’Shea J. J., Genome Biol., 2004, 5(12), 253 |
| [4] | Pesu M., Laurence A., Kishore N., Zwillich S. H., Chan G., O’Shea J. J., Immunol. Rev., 2008, 223, 132—142 |
| [5] | Witthuhn B. A., Quelle F. W., Silvennoinen O., Yi T., Tang B., Miura O., Ihle J. N., Cell, 1993, 74(2), 227—236 |
| [6] | Firmbach-Kraft I., Byers M., Shows T., Dalla-Favera R., Krolewski J. J., Oncogene, 1990, 5(9), 1329—1336 |
| [7] | Harpur A. G., Andres A. C., Ziemiecki A., Aston R. R., Wilks A. F., Oncogene, 1992, 7(7), 1347—1353 |
| [8] | Rane S. G., Reddy E. P., Oncogene, 1994, 9(8), 2415—2423 |
| [9] | Mercier E., Lissalde-Lavigne G.R., Gris J. C., N. Engl. J. Med., 2007, 357, 1984—1985 |
| [10] | Tono C., Xu G., Toki T., Takahashi Y., Sasaki S., Terui K., Ito E., Leukemia, 2005, 19, 1843—1844 |
| [11] | Chen E., Beer P. A., Godfrey A. L., Ortmann C. A., Li J., Costa-Pereira A. P., Ingle C. E., Dermitzakis E. T., Campbell P. J., Green A. R., Cancer Cell, 2010, 18, 524—535 |
| [12] | Bandaranayake R. M., Ungureanu D., Shan Y., Shaw D. E., Silvennoinen O., Hubbard S. R., Nat. Struct. Mol. Biol., 2012, 19(8), 754—759 |
| [13] | Clark J. D., Flanagan M. E., Telliez J. B., J. Med. Chem., 2014, 57(12), 5023—5038 |
| [14] | Menet C. J., Rompaey L. V., Geney R., Prog. Med. Chem., 2013, 52, 153—223 |
| [15] | Zhou T., Georgeon S., Moser R., Moore D. J., Caflisch A., Hantschel O., Leukemia, 2014, 28(2): 404—407 |
| [16] | Kang C. M., Zhao X. H., Yu Y. Q., Lü Y. T., Chem. J. Chinese Universities, 2016, 35(3), 550—554 |
| (康从民, 赵绪浩, 于玉琪, 吕英涛.高等学校化学学报,2016, 35(3), 550—554) | |
| [17] | Wu Y. J., Cui Y. L., Zheng Q. C., Zhang H. X., Chem. J. Chinese Universities, 2014, 35(12), 2605—2611 |
| (吴云剑, 崔颖璐, 郑清川, 张红星.高等学校化学学报,2014, 35(12), 2605—2611) | |
| [18] | Li X. H., Zhao J. W., Teng H., Chem. J. Chinese Universities, 2010, 31(2), 374—378 |
| (李晓晖, 赵俊伟, 滕虎.高等学校化学学报,2010, 31(2), 374—378) | |
| [19] | Zhuang S. L., Wang H. F., Ding K. K., Wang J. Y., Pan L. M., Lu Y. L., Liu Q. J., Zhang C. L., Chemosphere, 2016, 144, 1050—1059 |
| [20] | Muzzioli E., Del Rio A., Rastelli G., Chem. Boil. Drug Des., 2011, 78(2), 252—259 |
| [21] | Berman H. M., Battistuz T., Bhat T. N., Bluhm W. F., Bourne P. E., Burkhardt K., Feng Z., Gilliland G. L., Iype L., Jain S., Fagan P., Marvin J., Padilla D., Ravichandran V., Schneider B., Thanki N., Weissig H., Westbrook J. D., Zardecki C., Acta Crystallogr D, 2002, 58, 899—907 |
| [22] | Maestro, Version 8.5, Schrödinger, LLC. Cambridge, 2008 |
| [23] | Friesner R. A., Banks J. L., Murphy R. B., Halgren T. A., Klicic J. J., Mainz D. T., Repasky M. P., Knoll E. H., Shelley M., Perry J. K., Shaw D. E., Francis P., Shenkin P. S., J. Med. Chem., 2004, 47(7), 1739—1749 |
| [24] | Halgren T. A., Murphy R. B., Friesner R. A., Beard H. S., Frye L. L., Pollard W. T., Banks J. L., J. Med. Chem., 2004, 47(7), 1750—1759 |
| [25] | Case D. A., Cheatham T. E., Darden T., Gohlke H., Luo R., Merz K. M., Onufriev A., Simmerling C., Wang B.,Woods R. J., J. Comput. Chem., 2005, 26(16), 1668—1688 |
| [26] | Frisch M.J., Trucks G. W., Schlegel H. B., Scuseria G. E., Robb M. A., Cheeseman J. R., Scalmani G., Barone V., Mennucci B., Petersson G. A., Nakatsuji H., Caricato M., Li X., Hratchian H. P., Izmaylov A. F., Bloino J., Zheng G., Sonnenberg J. L., Hada M., Ehara M., Toyota K., Fukuda R., Hasegawa J., Ishida M., Nakajima T., Honda Y., Kitao O., Nakai H., Vreven T., Montgomery J. A. Jr., Peralta J. E., Ogliaro F., Bearpark M., Heyd J. J., Brothers E., Kudin K. N., Staroverov V. N., Kobayashi R., Normand J., Raghavachari K., Rendell A., Burant J. C., Iyengar S. S., Tomasi J., Cossi M., Rega N., Millam N. J., Klene M., Knox J. E., Cross J. B., Bakken V., Adamo C., Jaramillo J., Gomperts R., Stratmann R. E., Yazyev O., Austin A. J., Cammi R., Pomelli C., Ochterski J. W., Martin R. L., Morokuma K., Zakrzewski V. G., Voth G. A., Salvador P., Dannenberg J. J., Dapprich S., Daniels A. D., Farkas Ö., Foresman J. B., Ortiz J. V., Cioslowski J., Fox D. J., Gaussian 09, Revision A. 1, Gaussian Inc., Wallingford CT, 2009 |
| [27] | Wang J. M., Wang W., Kollman P. A., Case D. A., J. Mol. Graph. Model., 2006, 25(2), 247—260 |
| [28] | Paschek D., Day R., Garcia A. E., Phys. Chem. Chem. Phys., 2011, 13(44), 19840—19847 |
| [29] | Hou T. J., Wang J. M., Li Y. Y., Wang W., J. Chem. Inf. Model., 2011, 51(1), 69—82 |
| [1] | 宋有为, 安江伟, 王征, 王旭慧, 权燕红, 任军, 赵金仙. Ag,Zn,Pd掺杂对铜基催化剂草酸二甲酯选择性加氢反应的影响[J]. 高等学校化学学报, 2022, 43(6): 20210842. |
| [2] | 宋颖颖, 黄琳, 李庆森, 陈立妙. CuO/BiVO4光催化剂的制备及光催化CO2还原性能[J]. 高等学校化学学报, 2022, 43(6): 20220126. |
| [3] | 高志伟, 李军委, 史赛, 付强, 贾钧儒, 安海龙. 基于分子动力学模拟的TRPM8通道门控特性分析[J]. 高等学校化学学报, 2022, 43(6): 20220080. |
| [4] | 曾晛阳, 赵熹, 黄旭日. 细胞松弛素B对葡萄糖/质子共转运蛋白GlcPSe的抑制机理[J]. 高等学校化学学报, 2022, 43(4): 20210822. |
| [5] | 刘嘉欣, 闵杰, 许华杰, 任海生, 谈宁馨. 基于反应力场分子模拟的乙烯燃烧自由基与氮气相互作用研究[J]. 高等学校化学学报, 2022, 43(4): 20210834. |
| [6] | 陈瀚翔, 边绍菊, 胡斌, 李武. LiCl-NaCl-KCl-H2O溶液体系渗透压的分子动力学模拟[J]. 高等学校化学学报, 2022, 43(3): 20210727. |
| [7] | 胡波, 朱昊辰. 双层氧化石墨烯纳米体系中受限水的介电常数[J]. 高等学校化学学报, 2022, 43(2): 20210614. |
| [8] | 魏李娜, 彭莉, 朱锋, 顾鹏飞, 顾学红. 中空纤维Au-CeZr/FAU催化膜的制备及在富氢气氛CO选择性氧化反应中的应用[J]. 高等学校化学学报, 2022, 43(10): 20220175. |
| [9] | 李学宇, 王朝, 陈雅, 李可可, 李建全, 金顺敬, 陈丽华, 苏宝连. 等离激元共振光转热增强负载纳米金对丁二烯选择性加氢的催化性能[J]. 高等学校化学学报, 2022, 43(10): 20220174. |
| [10] | 张伶育, 张继龙, 曲泽星. RDX分子内振动能量重分配的动力学研究[J]. 高等学校化学学报, 2022, 43(10): 20220393. |
| [11] | 柳雪广, 杨晓珊, 马菁菁, 刘伟生. 铕基金属有机框架材料从混合染料中选择性分离亚甲基蓝[J]. 高等学校化学学报, 2022, 43(1): 20210715. |
| [12] | 李晨晨, 那永. 双功能复合材料g-C3N4/CdS/Ni催化光解水产氢和5-羟甲基糠醛氧化性能[J]. 高等学校化学学报, 2021, 42(9): 2896. |
| [13] | 李聪聪, 刘明皓, 韩佳睿, 朱镜璇, 韩葳葳, 李婉南. 基于分子动力学模拟的VmoLac非特异性底物催化活性的理论研究[J]. 高等学校化学学报, 2021, 42(8): 2518. |
| [14] | 雷晓彤, 金怡卿, 孟烜宇. 基于分子模拟方法预测PIP2在双孔钾通道TREK-1上结合位点的研究[J]. 高等学校化学学报, 2021, 42(8): 2550. |
| [15] | 史歌, 徐茜, 代枭, 张洁, 沈军, 宛新华. 芳香取代基结构对螺旋聚乙炔高效液相色谱手性固定相手性识别性能的影响[J]. 高等学校化学学报, 2021, 42(8): 2673. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||