

高等学校化学学报 ›› 2020, Vol. 41 ›› Issue (12): 2587.doi: 10.7503/cjcu20200592
• 庆祝《高等学校化学学报》复刊40周年专栏 • 上一篇 下一篇
李显明,郑婷,高露,李峰,侯贤灯,吴鹏
收稿日期:2020-08-23
出版日期:2020-12-10
发布日期:2020-12-09
作者简介:吴 鹏, 男, 博士, 研究员, 主要从事以室温磷光的光物理和光化学调控为基础, 探究其在分析化学领域的新应用研究.E-mail: 基金资助:
LI Xianming1, ZHENG Ting2, GAO Lu2, LI Feng1,2( ), HOU Xiandeng1,2, WU Peng1,2(
), HOU Xiandeng1,2, WU Peng1,2( )
)
Received:2020-08-23
Online:2020-12-10
Published:2020-12-09
Contact:
LI Feng
E-mail:windtalker_1205@scu.edu.cn;wupeng@scu.edu.cn
Supported by:摘要:
脱氧核糖核酸(DNA)放大技术对于核酸检测(NAT)至关重要. 聚合酶链式反应(PCR)虽然是核酸检测的基准扩增技术, 但其主要适用于条件较好的中心实验室. 重组酶聚合酶扩增(RPA)是一种非常有潜力的等温扩增技术, 对仪器设备依赖性小, 适用于资源贫乏地区. 因此, 该技术在核酸检测时不受实验场所限制, 非常适合即时检测(POCT). 作为一种正在快速发展的扩增技术, RPA也存在阻碍其进一步发展的缺陷. 本文对RPA的扩增原理和扩增性能进行了总结, 重点讨论了对扩增性能至关重要的引物重组和ATP动态平衡调控过程, 并概述了RPA存在的缺陷和潜在的解决方向.
中图分类号:
TrendMD:
李显明, 郑婷, 高露, 李峰, 侯贤灯, 吴鹏. 重组酶聚合酶扩增:扩增原理及性能分析. 高等学校化学学报, 2020, 41(12): 2587.
LI Xianming, ZHENG Ting, GAO Lu, LI Feng, HOU Xiandeng, WU Peng. Recombinase Polymerase Amplification: from Principle to Performance. Chem. J. Chinese Universities, 2020, 41(12): 2587.
| Category | Strategy I | Strategy II | Function description |
|---|---|---|---|
| Recombinase | uvsX | RecA | DNA?dependent ATPase, the key protein for DNA strand exchange |
ssDNA binding proteins | gp32 | SSB | Melting the secondary structure of primers to decrease undesirable interactions |
| Accessory proteins | uvsY | RecF, RecO | Assisting the loading of recombinase on primer |
| Crowding agents | PEGs, dectran, ficoll | Accelerating the RP process and primer extending | |
| Polymerase | T4 polymerase, Bsu DNA polymerase I, etc. | Extending the primer and displacement of the complementary strand of templates | |
| ATP regeneration | Phosphocreatine and creatine | Transforming ADP into ATP | |
| Reverse transcriptase | Mu?MLV reverse transcriptase | Converting RNA templates to ssDNA | |
Table 1 Functional substances involved in recombinase polymerase amplification
| Category | Strategy I | Strategy II | Function description |
|---|---|---|---|
| Recombinase | uvsX | RecA | DNA?dependent ATPase, the key protein for DNA strand exchange |
ssDNA binding proteins | gp32 | SSB | Melting the secondary structure of primers to decrease undesirable interactions |
| Accessory proteins | uvsY | RecF, RecO | Assisting the loading of recombinase on primer |
| Crowding agents | PEGs, dectran, ficoll | Accelerating the RP process and primer extending | |
| Polymerase | T4 polymerase, Bsu DNA polymerase I, etc. | Extending the primer and displacement of the complementary strand of templates | |
| ATP regeneration | Phosphocreatine and creatine | Transforming ADP into ATP | |
| Reverse transcriptase | Mu?MLV reverse transcriptase | Converting RNA templates to ssDNA | |
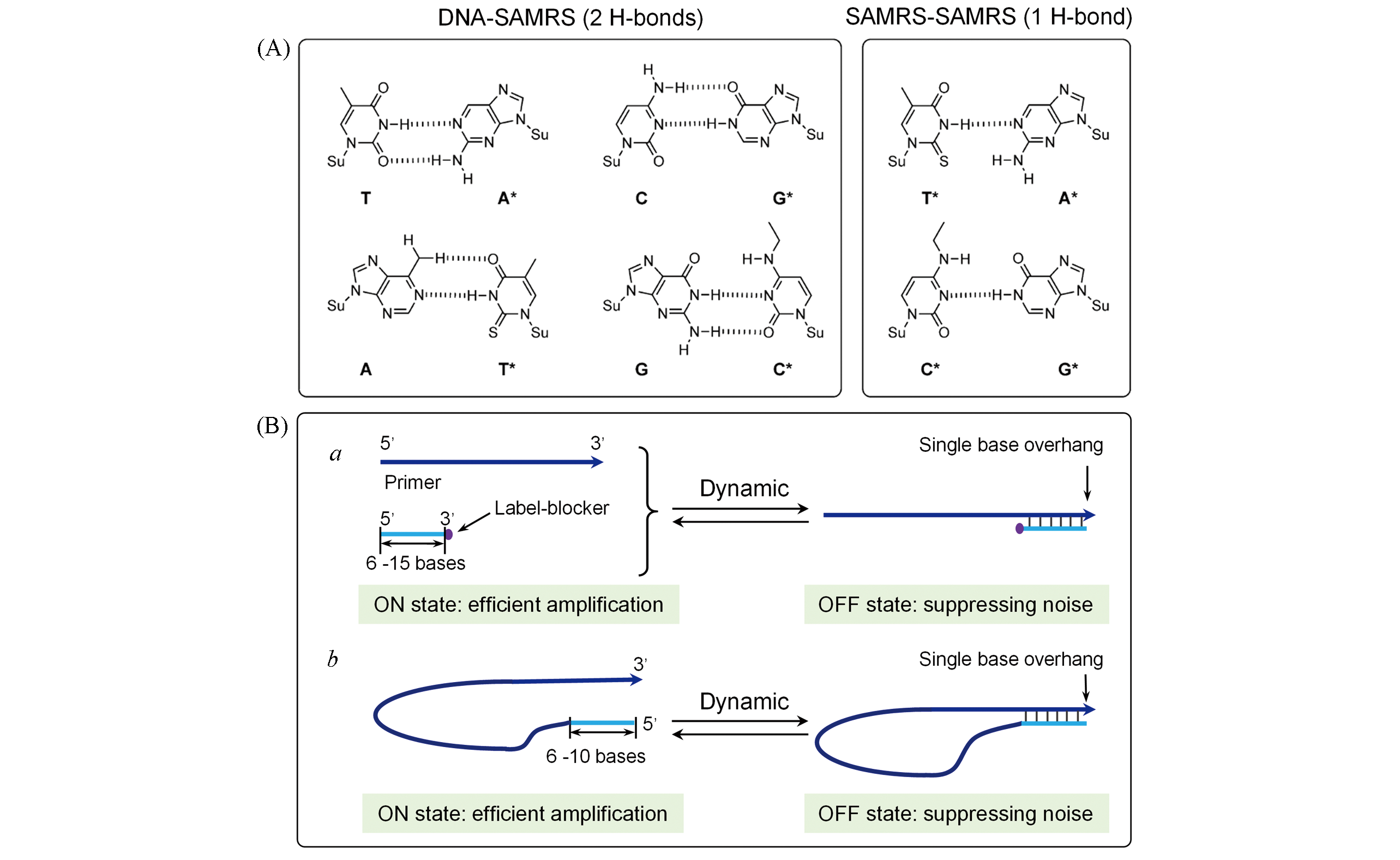
Fig.3 Primers for reducing nose products(A) Chemical formulas of the self?avoiding molecular?recognition system[33], Copyright 2014, Wiley?VCH;(B) competition designs.
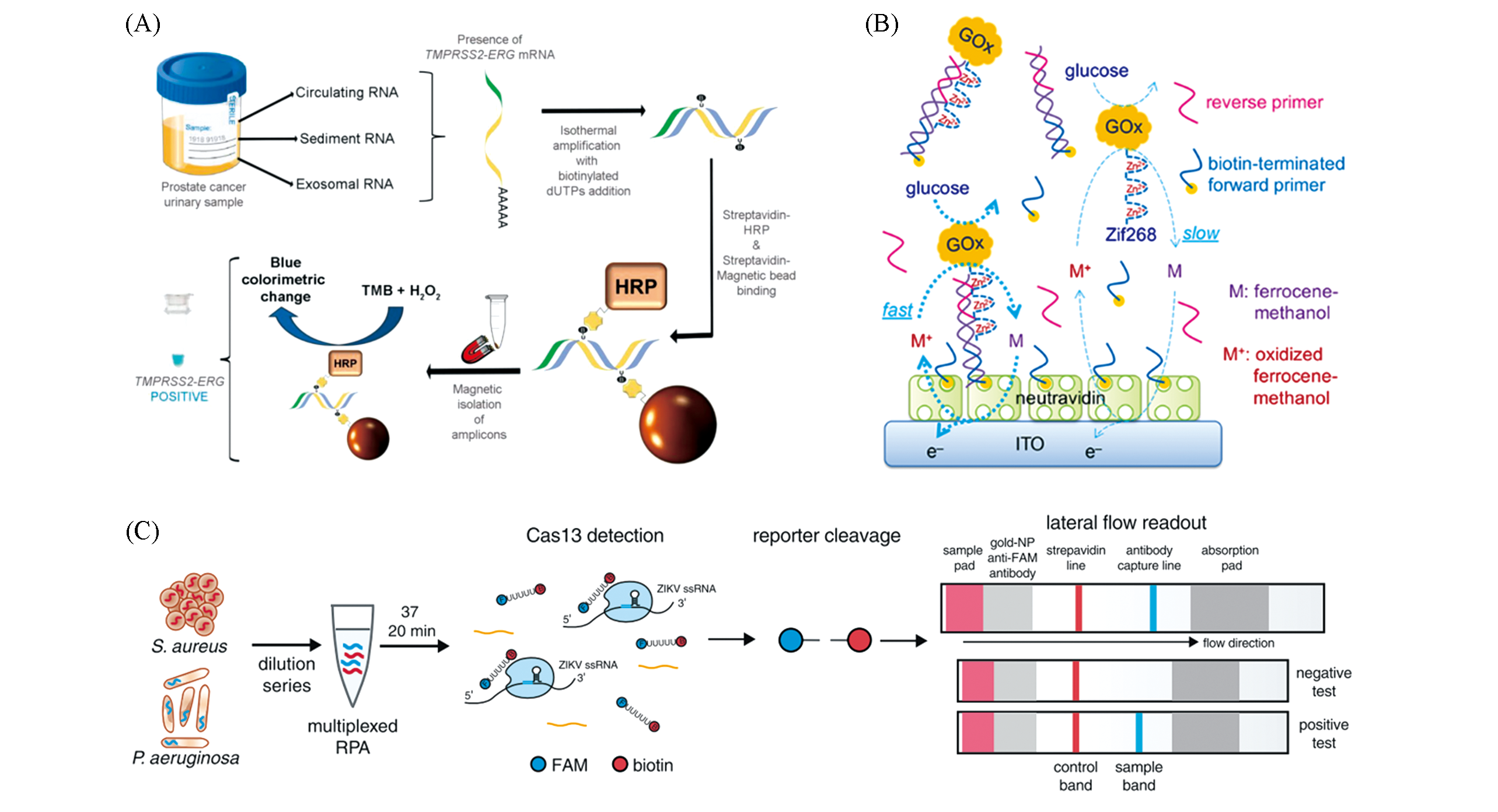
Fig.5 New amplicon testing methods(A) Colorimetric assay[45], Copyright 2018, Royal Society of Chemistry; (B) electrochemical method[46], Copyright 2018, Royal Society of Chemistry; (C) CTISPR/Cas based lateral flow readout[48], Copyright 2018 American Association for the Advancement of Science.
| Assaya | Primer number | Temperature/℃ | Reaction/min | Detection formatsb | Target | Reagent format | Multiplex | Ref. |
|---|---|---|---|---|---|---|---|---|
| NASBA | 2 | 41 | 105 | RTF, NALF | RNA(DNA) | Liquid | Y | [ |
| TMA | 2 | 60 | 140 | RTF | RNA(DNA) | Liquid | Y | [ |
| SMART | 2 | 41 | 140 | RTF | RNA, DNA | Liquid | N/A | [ |
| LAMP | 4 | 60—65 | 60—90 | RTF, NALF, RTT, TE | RNA, DNA | Liquid | N/A | [ |
| CPA | 4 | 65 | 65 | RTF, NALF | DNA | Liquid | N/A | [ |
| RCA | 1, 2 | 65 | 60 | RTF | RNA, DNA | Liquid | N/A | [ |
| RAM | 1 | 63 | 120—180 | RTF | RNA, DNA | Liquid | N/A | [ |
| SDA | 2 | 37 | 120 | RTF, NALF | RNA, DNA | Liquid | Y | [ |
| NEAR | 4 | 55 | 10 | RTF, NALF | RNA, DNA | Liquid | Y | [ |
| NEMA | 4 | 65 | 30 | NALF | DNA | Liquid | N/A | [ |
| ICA | 2 | 60 | 60 | RTF | DNA | Liquid | N/A | [ |
| EXPAR | 1 | 55 | 10—20 | RTF, NALF | DNA | Liquid | Y | [ |
| HDA | 2 | 65 | 75—90 | RTF, NALF | RNA, DNA | Liquid | Y | [ |
| RPA | 2 | 30—42 | 20 | RTF, NALF | RNA, DNA | Liquid, lyophilized | Y | [ |
Table 2 Comparison of current isothermal amplification methods
| Assaya | Primer number | Temperature/℃ | Reaction/min | Detection formatsb | Target | Reagent format | Multiplex | Ref. |
|---|---|---|---|---|---|---|---|---|
| NASBA | 2 | 41 | 105 | RTF, NALF | RNA(DNA) | Liquid | Y | [ |
| TMA | 2 | 60 | 140 | RTF | RNA(DNA) | Liquid | Y | [ |
| SMART | 2 | 41 | 140 | RTF | RNA, DNA | Liquid | N/A | [ |
| LAMP | 4 | 60—65 | 60—90 | RTF, NALF, RTT, TE | RNA, DNA | Liquid | N/A | [ |
| CPA | 4 | 65 | 65 | RTF, NALF | DNA | Liquid | N/A | [ |
| RCA | 1, 2 | 65 | 60 | RTF | RNA, DNA | Liquid | N/A | [ |
| RAM | 1 | 63 | 120—180 | RTF | RNA, DNA | Liquid | N/A | [ |
| SDA | 2 | 37 | 120 | RTF, NALF | RNA, DNA | Liquid | Y | [ |
| NEAR | 4 | 55 | 10 | RTF, NALF | RNA, DNA | Liquid | Y | [ |
| NEMA | 4 | 65 | 30 | NALF | DNA | Liquid | N/A | [ |
| ICA | 2 | 60 | 60 | RTF | DNA | Liquid | N/A | [ |
| EXPAR | 1 | 55 | 10—20 | RTF, NALF | DNA | Liquid | Y | [ |
| HDA | 2 | 65 | 75—90 | RTF, NALF | RNA, DNA | Liquid | Y | [ |
| RPA | 2 | 30—42 | 20 | RTF, NALF | RNA, DNA | Liquid, lyophilized | Y | [ |
| 1 | Wang R. F., Cao W. W., Cerniglia C. E., J. Appl. Microbiol., 1997, 83(6), 727—736 |
| 2 | Bustin Stephen A., Mueller R., Clin. Sci., 2005, 109(4), 365—379 |
| 3 | Morling N., Biochem. Soc. Trans., 2009, 37(2), 438—440 |
| 4 | Clerc O., Greub G., Clin. Microbiol. Infect., 2010, 16(8), 1054—1061 |
| 5 | Dong T. Y., Mansour H., Hu H., Wang G. A., Watson C. J. F., Yousef M., Matamoros G., Sanchez A. L., MacNeil A. J., Wu P., Li F., Anal. Chem., 2020, 92(9), 6456—6461 |
| 6 | Daher R. K., Stewart G., Boissinot M., Bergeron M. G., Clin. Chem., 2016, 947—958 |
| 7 | Lobato I. M., O’Sullivan C. K., Trends Anal. Chem., 2017, 19—35 |
| 8 | Li J., Macdonald J., von Stetten F., Analyst, 2019, 144(1), 31—67 |
| 9 | Stringer O. W., Andrews J. M., Greetham H. L., Forrest M. S., Nat. Methods, 2018, 15(5), I—III |
| 10 | Piepenburg O., Williams C. H., Stemple D. L., Armes N. A., PLoS Biol., 2006, 4(7), e204 |
| 11 | Zaghloul H., El⁃shahat M., World J. Hepatol., 2014, 6(12), 916—922 |
| 12 | James A., Macdonald J., Expert Rev. Mol. Diagn., 2015, 15(11), 1475—1489 |
| 13 | Moore M. D., Jaykus L. A., Future Virol., 2017, 12(8), 421—429 |
| 14 | Babu B., Ochoa⁃Corona F. M., Paret M. L., Anal. Biochem., 2018, 546, 72—77 |
| 15 | Piepenburg O., Williams Colin H., Armes Niall A., Stemple Derek L., Recombinase Polymerase Amplification, US 20190360030, 2019⁃11⁃28 |
| 16 | Kowalczykowski S. C., Krupp R. A., J. Mol. Biol., 1987, 193(1), 97—113 |
| 17 | Bar⁃Ziv R., Libchaber A., Proc. Natl. Acad. Sci. USA, 2001, 98(16), 9068—9073 |
| 18 | Roy R., Kozlov A. G., Lohman T. M., Ha T., Nature, 2009, 461(7267), 1092—1097 |
| 19 | Liu J., Morrical S. W., Virol J., 2010, 7(1), 357 |
| 20 | Mok E., Wee E., Wang Y., Trau M., Sci. Rep., 2016, 6(1), 37837 |
| 21 | Bianco P. R., Tracy R. B., Kowalczykowski S. C., Front. Biosci., 1998, 3, 570—603 |
| 22 | Bell J. C., Kowalczykowski S. C., Trends Biochem. Sci., 2016, 41(6), 491—507 |
| 23 | Kowalezykowski S. C., Annu. Rev. Biophys. Biophys. Chem., 1991, 20(1), 539—575 |
| 24 | Rosselli W., Stasiak A., EMBO J., 1991, 10(13), 4391—4396 |
| 25 | Danilowicz C., Hermans L., Coljee V., Prévost C., Prentiss M., Nucleic Acids Res., 2017, 45(14), 8448—8462 |
| 26 | Lee J. Y., Qi Z., Greene E. C., J. Biol. Chem., 2016, 291(42), 22218—22230 |
| 27 | Bittl J. A., Ingwall J. S., J. Biol. Chem., 1985, 260(6), 3512—3517 |
| 28 | Zhao B., Zhang D., Li C., Yuan Z., Yu F., Zhong S., Jiang G., Yang Y. G., Le X. C., Weinfeld M., Zhu P., Wang H., Cell Discov., 2017, 3(1), 16053 |
| 29 | Kim S. H., Ragunathan K., Park J., Joo C., Kim D., Ha T., J. Am. Chem. Soc., 2014, 136(42), 14796—14800 |
| 30 | Morrical S. W., Alberts B. M., J. Biol. Chem., 1990, 265(25), 15096—15103 |
| 31 | Cox M. M., Crit. Rev. Biochem. Mol. Biol., 2007, 42(1), 41—63 |
| 32 | Cox M. M., The Bacterial RecA Protein: Structure, Function, and Regulation, Springer Berlin Heidelberg, Berlin, 2007, 53—94 |
| 33 | Sharma N., Hoshika S., Hutter D., Bradley K. M., Benner S. A., ChemBioChem, 2014, 15(15), 2268—2274 |
| 34 | Higgins M., Ravenhall M., Ward D., Phelan J., Ibrahim A., Forrest M. S., Clark T. G., Campino S., Bioinformatics, 2018, 35(4), 682—684 |
| 35 | del Río J. S., Adly N. Y., Acero⁃Sánchez J. L., Henry O. Y., O’Sullivan C. K., Biosens. Bioelectron., 2014, 54, 674—678 |
| 36 | Crannell Z. A., Cabada M. M., Castellanos⁃Gonzalez A., Irani A., White A. C., Richards⁃Kortum R., Am. J. Trop. Med. Hyg., 2015, 92(3), 583—587 |
| 37 | Kunze A., Dilcher M., Abd El Wahed A., Hufert F., Niessner R., Seidel M., Anal. Chem., 2015, 88(1), 898—905 |
| 38 | Ng B. Y., Wee E. J., West N. P., Trau M., Sci. Rep., 2015, 5, 15027 |
| 39 | Kim J., Biondi M. J., Feld J. J., Chan W. C., ACS Nano, 2016, 10(4), 4742—4753 |
| 40 | Liu H. B., Du X. J., Zang Y. X. Li P., Wang S., J. Agric. Food Chem., 2017, 65(47), 10290—10299 |
| 41 | Chertow D. S., Science, 2018, 360(6387), 381—382 |
| 42 | Shin Y., Perera A. P., Kim K. W., Park M. K., Lab Chip, 2013, 13(11), 2106—2114 |
| 43 | Lau H. Y., Wang Y., Wee E. J., Botella J. R., Trau M., Anal. Chem., 2016, 88(16), 8074—8081 |
| 44 | Ng B. Y., Wee E. J., West N. P., Trau M., ACS Sens., 2015, 1(2), 173—178 |
| 45 | Islam M. N., Moriam S., Umer M., Phan H. P., Salomon C., Kline R., Nguyen N. T., Shiddiky M. J., Analyst, 2018, 3021—3028 |
| 46 | Fang C. S., Kim K. S., Ha D. T., Kim M. S., Yang H., Anal. Chem., 2018, 90(7), 4776—4782 |
| 47 | Gootenberg J. S., Abudayyeh O. O., Lee J. W., Essletzbichler P., Dy A. J., Joung J., Verdine V., Donghia N., Daringer N. M., Freije C. A., Science, 2017, 356(6336), 438—442 |
| 48 | Gootenberg J. S., Abudayyeh O. O., Kellner M. J., Joung J., Collins J. J., Zhang F., Science, 2018, 360(6387), 439—444 |
| 49 | Chen J. S., Ma E., Harrington L. B., Da Costa M., Tian X., Palefsky J. M., Doudna J. A., Science, 2018, 360(6387), 436—439 |
| 50 | Myhrvold C., Freije C. A., Gootenberg J. S., Abudayyeh O. O., Metsky H. C., Durbin A. F., Kellner M. J., Tan A. L., Paul L. M., Parham L. A., Science, 2018, 360(6387), 444—448 |
| 51 | Kunze A., Dilcher M., Abd El Wahed A., Hufert F., Niessner R., Seidel M., Anal. Chem., 2015, 88(1), 898—905 |
| 52 | Kober C., Niessner R., Seidel M.,. Biosens. Bioelectron., 2018, 100, 49—55 |
| 53 | Elsäßer D., Ho J., Niessner R., Tiehm A., Seidel M., Anal. Biochem., 2018, 546, 58—64 |
| 54 | Lau H. Y., Wang Y., Wee E. J., Botella J. R., Trau M., Anal. Chem., 2016, 88(16), 8074—8081 |
| 55 | Koo K. M. Wee E. J., Mainwaring P. N., Wang Y., Trau M., Small, 2016, 12(45), 6233—6242 |
| 56 | Gracias K. S., Mckillip J. L., J. Rapid Methods Autom. Microbiol., 2007, 15(3), 295—309 |
| 57 | Hofmann W. P., Dries V., Herrmann E., Gärtner B., Zeuzem S., Sarrazin C., J. Clin. Virol., 2005, 32(4), 289—293 |
| 58 | Wharam S. D., Hall M. J., Wilson W. H., Virol. J., 2007, 4(1), 52 |
| 59 | Curtis K. A., Rudolph D. L., Owen S. M., J. Med. Virol., 2009, 81(6), 966—972 |
| 60 | Mori Y., Notomi T., J. Infect. Chemother., 2009, 15(2), 62—69 |
| 61 | Fang R., Li X., Hu L., You Q., Li J., Wu J., Xu P., Zhong H., Luo Y., Mei J., Gao Q., J. Clin. Microbiol., 2009, 47(3), 845—847 |
| 62 | Johne R., Müller H., Rector A., van Ranst M., Stevens H., Trends Microbiol., 2009, 17(5), 205—211 |
| 63 | Yao B., Li J., Huang H., Sun C., Wang Z., Fan Y., Chang Q., Li S., Xi J., RNA, 2009, 15(9), 1787—1794 |
| 64 | Hellyer T. J., Nadeau J. G., Expert Rev. Mol. Diagn., 2004, 4(2), 251—261 |
| 65 | McHugh T. D., Pope C. F., Ling C. L., Patel S., Billington O. J., Gosling R. D., Lipman M. C., Gillespie S. H., J. Med. Microbiol., 2004, 53(12), 1215—1219 |
| 66 | Maples B. K., Holmberg R. C., Miller A. P.,Provins J., Rot R., Mandell J., Nicking and Extension Amplification Reaction for the Exponential Amplification of Nucleic Acids, US 9562264, 2017⁃07⁃02 |
| 67 | You Q. M., Hu L., Wan J., Zhong H. Y., Method for Amplifying Target Nucleic Acid Sequence by Nickase, and Kit for Amplifying Target Nucleic Acid Sequence and Its Use, CP 1811447A, 2006⁃08⁃02 |
| 68 | Jung C., Chung J. W., Kim U. O., Kim M. H., Park H. G., Anal. Chem., 2010, 82(14), 5937—5943 |
| 69 | van Ness J., van Ness L. K., Galas D. J., Proc. Natl. Acad. Sci. USA, 2003, 100(8), 4504—4509 |
| 70 | Tan E., Erwin B., Dames S., Voelkerding K., Niemz A., Clin. Chem., 2007, 53(11), 2017—2020 |
| 71 | Jeong Y. J., Park K., Kim D. E., Cell. Mol. Life Sci., 2009, 66(20), 3325 |
| 72 | Vincent M., Xu Y., Kong H., EMBO Rep., 2004, 5(8), 795—800 |
| 73 | Lutz S., Weber P., Focke M., Faltin B., Hoffmann J., Müller C., Mark D., Roth G., Munday P., Armes N., Piepenburg O., Zengerle R., von Stetten F., Lab Chip, 2010, 10(7), 887—893 |
| 74 | Koo K. M., Wee E. J., Trau M., Theranostics, 2016, 6(9), 1415—1424 |
| 75 | Hu C., Kalsi S., Zeimpekis I., Sun K., Ashburn P., Turner C., Sutton J. M. Morgan H., Biosens. Bioelectron., 2017, 96, 281—287 |
| 76 | Koo K. M., Wee E. J., Trau M., Biosens. Bioelectron., 2017, 89, 715—720 |
| 77 | Ng B. Y., Wee E. J., Woods K., Anderson W., Antaw F., Tsang H. Z., West N. P., Trau M., Anal. Chem., 2017, 89(17), 9017—9022 |
| 78 | James A., Macdonald J., Expert Rev. Mol. Diagn., 2015, 15(11), 1475—1489 |
| 79 | Kissenkötter J., Böhlken⁃Fascher S., Forrest M. S., Piepenburg O., Czerny C. P., Abd El Wahed A., Food Chem., 2020, 322, 126759 |
| 80 | Kalsi S., Valiadi M., Tsaloglou M. N., Parry⁃Jones L., Jacobs A., Watson R., Turner C., Amos R., Hadwen B., Buse J., Lab Chip, 2015, 15(14), 3065—3075 |
| 81 | Yeh E. C., Fu C. C., Hu L., Thakur R., Feng J., Lee L. P., Sci. Adv., 2017, 3(3), e1501645 |
| 82 | Li J., Ma B., Fang J., Zhi A., Chen E., Xu Y., Yu X., Sun C., Zhang M., Foods, 2020, 9(1), 27 |
| 83 | Lutz S., Weber P., Focke M., Faltin B., Hoffmann J., Müller C., Mark D., Roth G., Munday P., Armes N., Piepenburg O., Zengerle R., von Stetten F., Lab Chip, 2010, 10(7), 887—893 |
| 84 | Foudeh A. M., Fatanat Didar T., Veres T., Tabrizian M., Lab Chip, 2012, 12(18), 3249—3266 |
| 85 | Kim T. H., Park J., Kim C. J., Cho Y. K., Anal. Chem., 2014, 86(8), 3841—3848 |
| 86 | Chen J., Xu Y., Yan H., Zhu Y., Wang L., Zhang Y., Lu Y., Xing W., Lab Chip, 2018, 18(16), 2441—2452 |
| 87 | Rohrman B. A., Richards⁃Kortum R. R., Lab Chip, 2012, 12(17), 3082—3088 |
| 88 | Ahn H., Batule B. S., Seok Y., Kim M. G., Anal. Chem., 2018, 90(17), 10211—10216 |
| 89 | Ming K., Kim J., Biondi M. J., Syed A., Chen K., Lam A., Ostrowski M., Rebbapragada A., Feld J. J., Chan W. C., ACS Nano, 2015, 9(3), 3060—3074 |
| 90 | Amaratunga M., Benight A. S., Biochem. Biophys. Res. Commun., 1988, 157(1), 127—133 |
| 91 | Kim S. H., Joo C., Ha T., Kim D., Nucleic Acids Res., 2013, 41(16), 7738—7744 |
| 92 | Wang Y., Zhou R., Liu W., Liu C., Wu P., Chin. Chem. Lett., 2020, doi: 10.1016/j.cclet.2020.01.023 |
| 93 | Butler B. C., Hanchett R. H., Rafailov H., MacDonald G., Biophys. J., 2002, 82(4), 2198—2210 |
| 94 | Lenhart J. S., Brandes E. R., Schroeder J. W., Sorenson R. J., Showalter H. D., Simmons L. A., J. Bacteriol., 2014, 196(15), 2851—2860 |
| 95 | Drain P. K., Hyle E. P., Noubary F., Freedberg K. A., Wilson D., Bishai W. R., Rodriguez W., Bassett I. V., Lancet Infect. Dis., 2014, 14(3), 239—249 |
| [1] | 魏敏敏, 袁泽, 闾敏, 马辉, 谢小吉, 黄岭. 稀土掺杂上转换纳米颗粒-金属有机骨架复合材料的研究进展[J]. 高等学校化学学报, 2021, 42(8): 2313. |
| [2] | 葛浩英, 杜健军, 龙飒然, 孙文, 樊江莉, 彭孝军. 功能化金纳米材料在肿瘤诊疗中的研究与应用[J]. 高等学校化学学报, 2021, 42(4): 1202. |
| [3] | 胡灵, 殷垚, 柯国梁, 张晓兵. 基于DNA纳米结构的细胞间相互作用的调控[J]. 高等学校化学学报, 2021, 42(11): 3284. |
| [4] | 王庆, 何雨秋, 王富安. 多功能脱氧核酶用于生物医学分析的研究进展[J]. 高等学校化学学报, 2021, 42(11): 3334. |
| [5] | 席京, 陈娜, 杨雁冰, 袁荃. 长余辉纳米材料的控制合成及在疾病诊断中的应用[J]. 高等学校化学学报, 2021, 42(11): 3247. |
| [6] | 刘园, 邓瑾琦, 赵帅, 田飞, 李轶, 孙佳姝, 刘超. 基于分子识别的免疫层析技术用于新冠肺炎感染的快速诊断[J]. 高等学校化学学报, 2021, 42(11): 3390. |
| [7] | 罗成, 彭雅梅, 沈宏, 方群, 潘建章. 多指标免疫分析的研究进展[J]. 高等学校化学学报, 2021, 42(11): 3421. |
| [8] | 彭伙, 高则航, 廖承悦, 王晓冬, 周洪波, 赵建龙. 一种用于核酸绝对定量检测的高鲁棒性液滴式数字PCR芯片[J]. 高等学校化学学报, 2020, 41(8): 1760. |
| [9] | 董倩, 李兆倩, 彭天欢, 陈卓, 谭蔚泓. 核酸适体在癌症诊疗中的研究进展[J]. 高等学校化学学报, 2020, 41(12): 2648. |
| [10] | 石越, 毛庆, 肖成, 景维云, 张学元. PtRu/C表面甲醇电催化氧化动力学的非线性谱学分析[J]. 高等学校化学学报, 2018, 39(9): 2017. |
| [11] | 康晓燕, 贺安琪, 王泾丹, 郭然, 翟延君, 徐怡庄, 吴瑾光. 碳酸镧分离富集DNA的研究[J]. 高等学校化学学报, 2016, 37(1): 7. |
| [12] | 马立娜, 柳扶摇, 高嘉雪, 王振新. 基于聚多巴胺纳米粒子的荧光共振能量转移法检测microRNA[J]. 高等学校化学学报, 2015, 36(6): 1061. |
| [13] | 陈杰, 李小舟, 田华雨, 朱筱娟, 陈学思. 两性离子聚合物在恶性肿瘤治疗中的应用[J]. 高等学校化学学报, 2015, 36(11): 2148. |
| [14] | 李敏, 邢朝晖, 田艳芝, 姜勇. CMC-g-DNA接枝共聚物的合成与表征[J]. 高等学校化学学报, 2014, 35(7): 1590. |
| [15] | 于永江, 魏光耀, 周颖, 李文超, 李秋顺, 王珺楠, 姚卫国, 董文飞. 聚电解质结构及盐浓度对基于DNA和聚阳离子的层层组装型大孔薄膜结构的影响[J]. 高等学校化学学报, 2013, 34(11): 2644. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||