

高等学校化学学报 ›› 2020, Vol. 41 ›› Issue (11): 2345.doi: 10.7503/cjcu20200397
• 庆祝《高等学校化学学报》复刊40周年专栏 • 上一篇 下一篇
收稿日期:2020-06-29
出版日期:2020-11-10
发布日期:2020-11-06
通讯作者:
顾宏周
E-mail:hongzhou.gu@fudan.edu.cn
基金资助:
TONG Zongxuan, HU Qinqin, GU Hongzhou( )
)
Received:2020-06-29
Online:2020-11-10
Published:2020-11-06
Contact:
GU Hongzhou
E-mail:hongzhou.gu@fudan.edu.cn
Supported by:摘要:
综述了脱氧核糖核酸酶(DNA酶)的起源及分离富集策略, 对比了DNA酶与核糖核酸酶(RNA酶)及蛋白酶的相似点和不同之处, 并重点讨论了产物捕获和对冲抵消等策略对筛选获得DNA酶的独到之处; 同时系统回顾了近年来分离出的可特异感应各种金属离子或生物样本(包括细菌、 细胞等), 从而能在特定位点切割RNA底物的DNA酶探针; 阐述了DNA酶领域现存的挑战, 总结和展望了新思路和新方向.
中图分类号:
TrendMD:
佟宗轩, 胡沁沁, 顾宏周. DNA酶: 筛选、 生物传感及展望. 高等学校化学学报, 2020, 41(11): 2345.
TONG Zongxuan, HU Qinqin, GU Hongzhou. Deoxyribozymes: Selection, Biosensing and Outlook. Chem. J. Chinese Universities, 2020, 41(11): 2345.
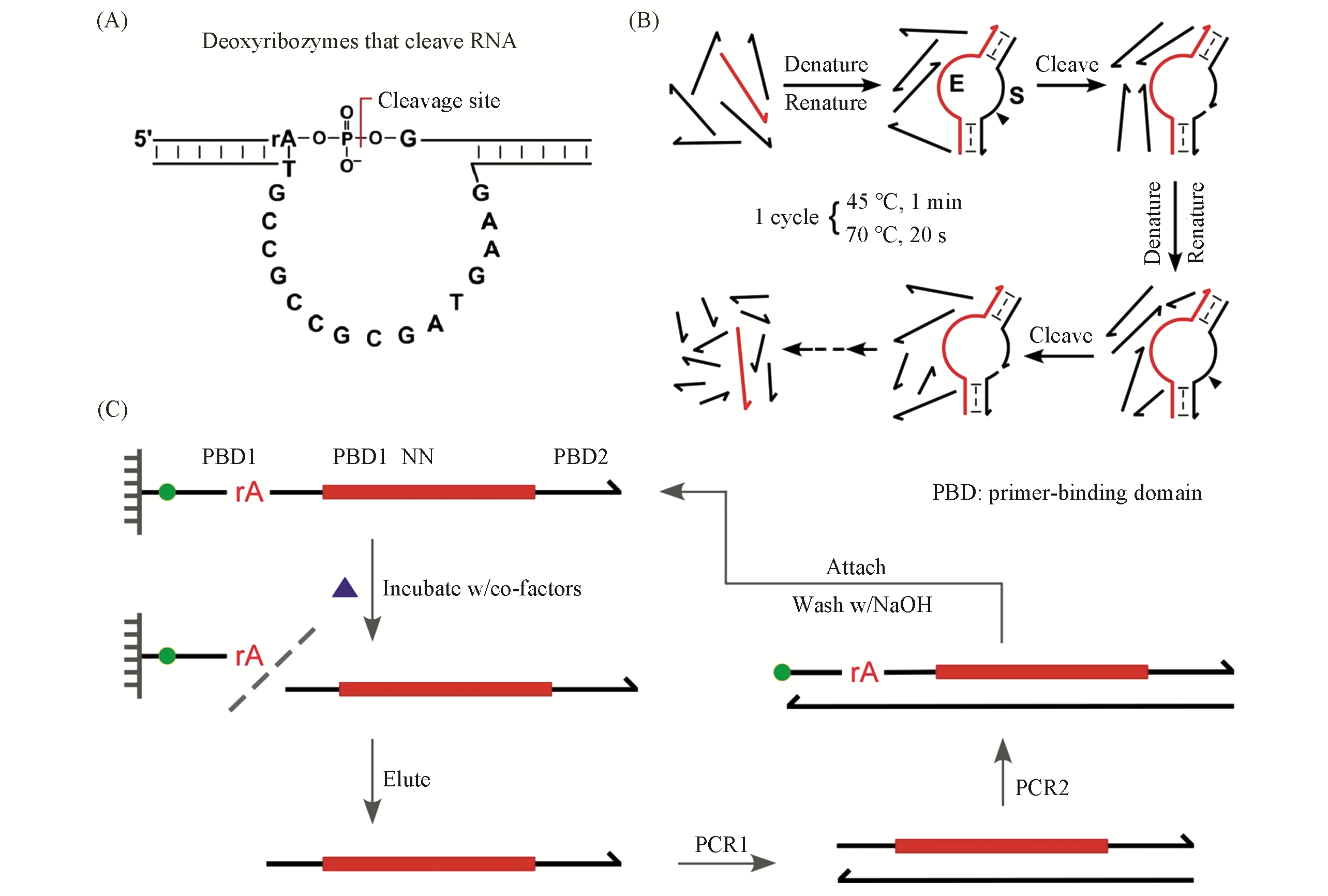
Fig.1 Deoxyribozymes and the classical selection strategy to isolate them(A) The first deoxyribozymes that cleave RNA; (B) thermal-cycling strategy to promote deoxyribozymes’ multiple- turnover rate[16], Copyright 2018, American Chemical Society; (C) the classical selection strategy to isolate RNA-cleaving deoxyribozymes.
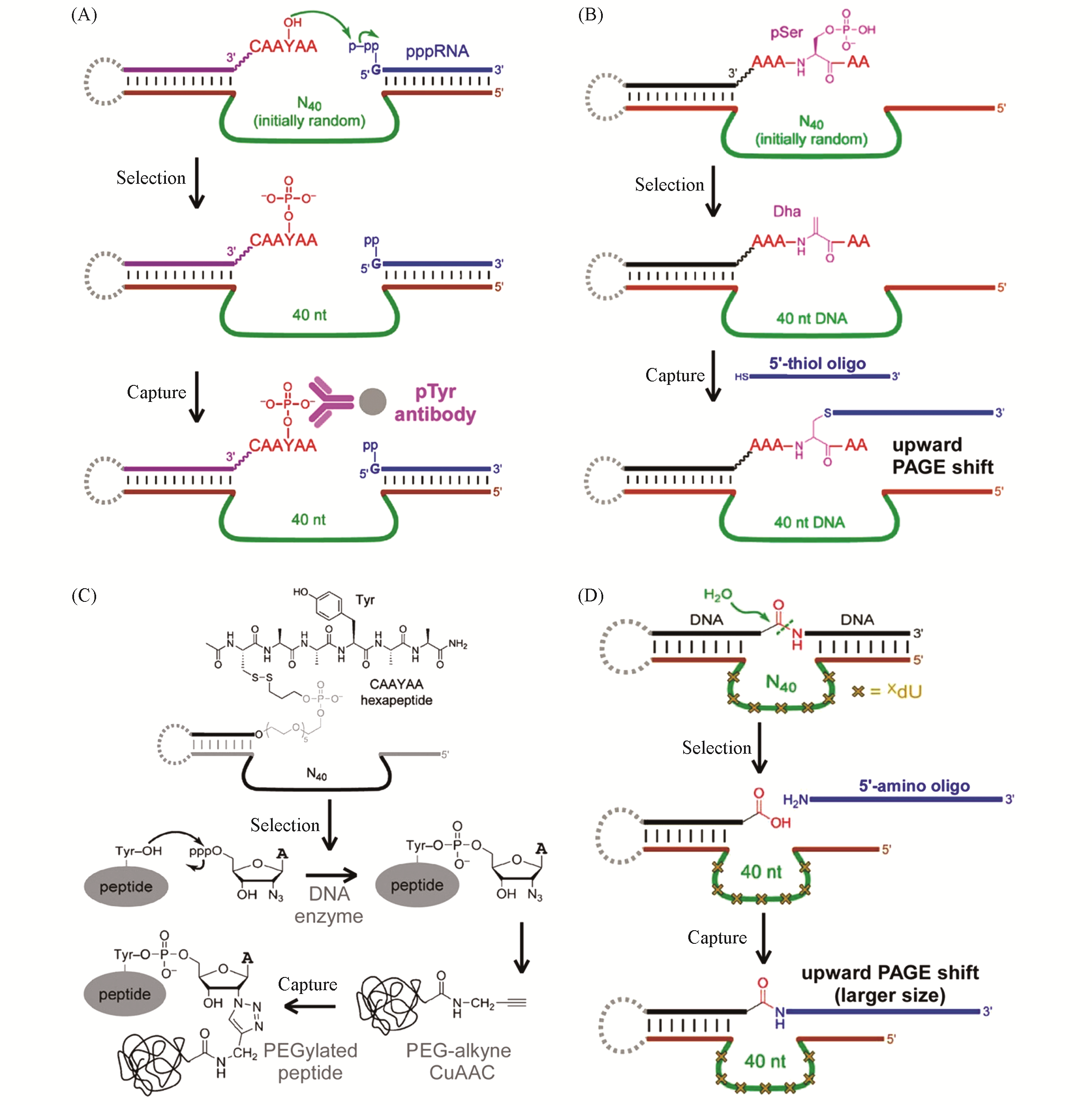
Fig.2 The product?capturing strategy to isolate deoxyribozymes that catalyze reactions on proteins/peptides(A) Phosphorylation of Tyr[26], Copyright 2013, American Chemical Society; (B) phosphorylated Ser to Dha[28], Copyright 2015, American Chemical Society; (C) N3-modification on Tyr[29], Copyright 2016, Wiley-VCH; (D) hydrolysis of peptide linkage[31], Copyright 2016, American Chemical Society.
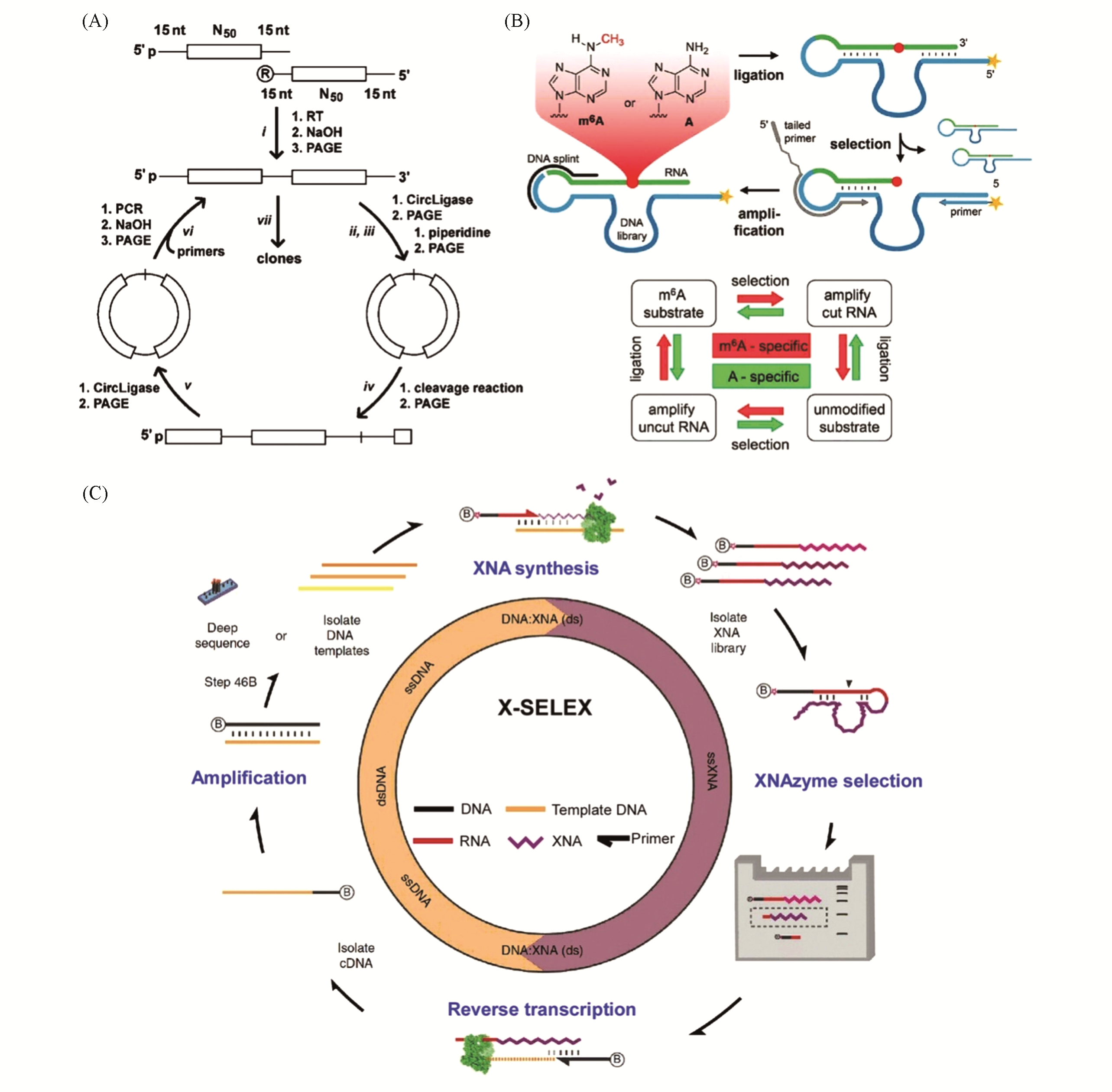
Fig.3 Strategies to isolate deoxyribozymes that catalyze reactions on nucleic acids(A) Hydrolysis of DNA[32], Copyright 2013, American Chemical Society; (B) m6A-responding RNA cleavage[35], Copyright 2018, Wiley-VCH; (C) X-SELEX for the isolation of XNAzymes[38], Copyright 2015, Springer Nature.
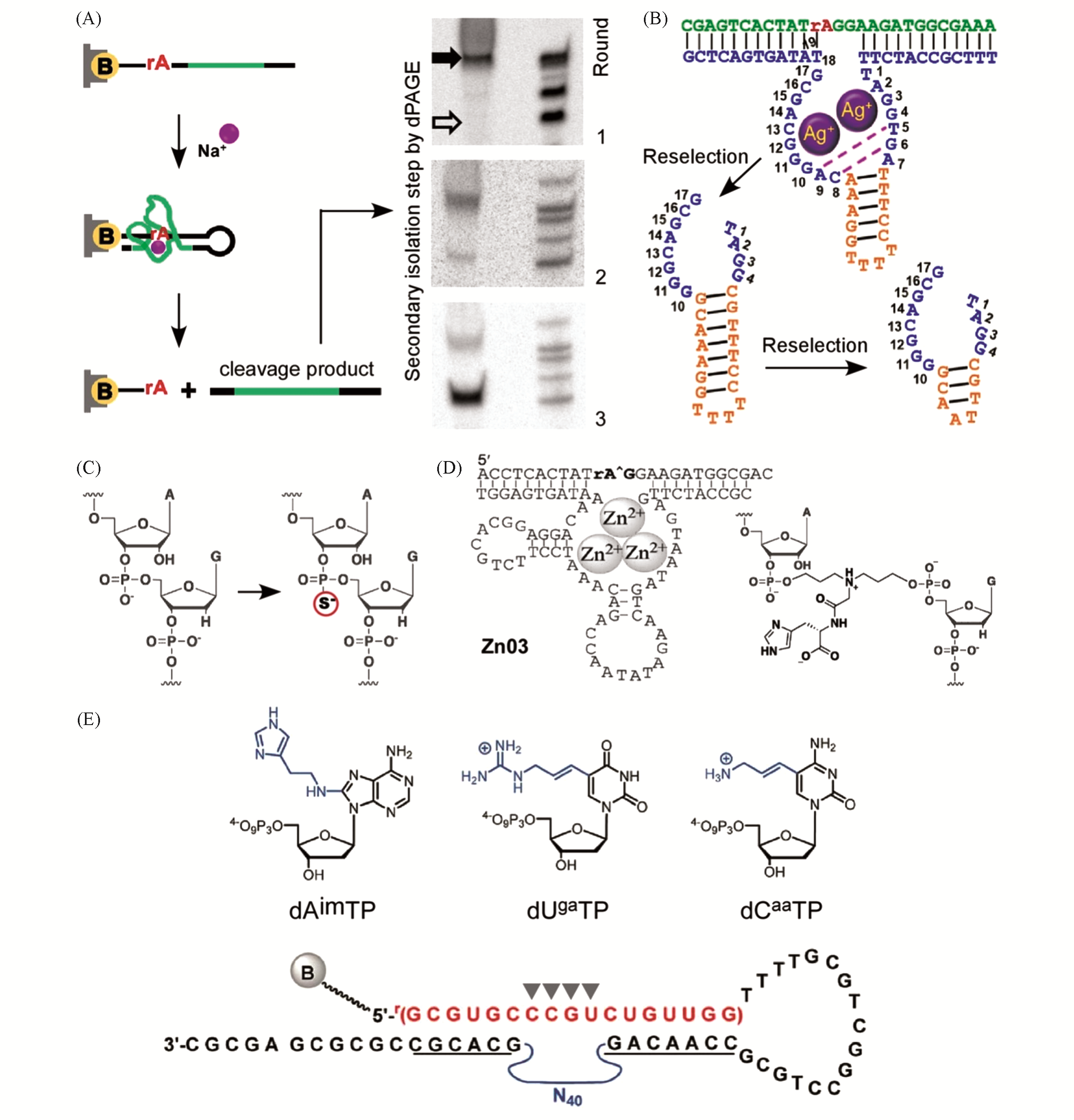
Fig.4 Optimizing strategies to select the metal?ion?dependent RNA?cleaving deoxyribozymes(A) Na+-dependent[53], Copyright 2015, National Academy of Sciences; (B) Ag+-dependent[57], Copyright 2018, American Che- mical Society; (C) phosphor-to-sulfur substitution[58], Copyright 2016, American Chemical Society; (D) multi-Zn2+-dependent[60], Copyright 2020, Wiley-VCH; (E) base-modifications in the catalytic domain[61], Copyright 2018, The Royal Society of Chemistry.
| 1 | Breaker R. R., Joyce G. F., Chem. Biol., 1994, 1(4), 223—229 |
| 2 | Baum D. A., Silverman S. K., Cell. Mol. Life Sci., 2008, 65(14), 2156—2174 |
| 3 | Fokina A. A., Chelobanov B. P., Fujii M., Stetsenko D. A., Expert Opin. Drug Delivery, 2017, 14(9), 1077—1089 |
| 4 | Hollenstein M., Molecules, 2015, 20(9), 20777—20804 |
| 5 | Silverman S. K., Nucleic Acids Res., 2005, 33(19), 6151—6163 |
| 6 | Silverman S. K., Trends Biochem. Sci., 2016, 41(7), 595—609 |
| 7 | Ali M. M., Wolfe M., Tram K., Gu J., Filipe C. D. M., Li Y., Brennan J. D., Angew. Chem. Int. Ed., 2019, 49(51), 9907—9911 |
| 8 | Lake R. J., Yang Z., Zhang J., Lu Y., Acc. Chem. Res., 2019, 52(12), 3275—3286 |
| 9 | Wang H., Chen Y., Wang H., Liu X., Zhou X., Wang F., Angew. Chem. Int. Ed., 2019, 58(7), 7380—7384 |
| 10 | Meng L., Ma W., Lin S., Shi S., Li Y., Lin Y., ACS Appl. Mater. Interfaces,2019, 11(7), 6850—6857 |
| 11 | Feng J., Xu Z., Liu F., Zhao Y., Yu W., Pan M., Wang F., Liu X., ACS Nano, 2018, 12(4), 12888—12901 |
| 12 | Wu Z., Fan H., Satyavolu N. S. R., Wang W., Lake R., Jiang J. H., Lu Y., Angew. Chem. Int. Ed., 2017, 56(30), 8721—8725 |
| 13 | Liu C., Hu Y., Pan Q., Yi J., Zhang J., He M., He M., Chen T., Chu X., Biosens. Bioelectron., 2019,136(1), 31—37 |
| 14 | Ponce⁃Salvatierra A., Wawrzyniak⁃Turek K., Steuerwald U., Höbartner C., Pena V., Nature, 2016, 529(7585), 231—234 |
| 15 | Liu H., Yu X., Chen Y., Zhang J., Wu B., Zheng L., Haruehanroengra P., Wang R., Li S., Lin J., Li J., Sheng J., Huang Z., Ma J., Gan J., Nat. Commun., 2017, 8(1), 2006—2015 |
| 16 | Du X., Zhong X., Li W., Li H., Gu H., ACS Catal., 2018, 8(7), 5996—6005 |
| 17 | Tram K., Xia J., Gysbers R., Li Y., PloS One, 2015, 10(1), e0126402 |
| 18 | Schlosser K., Gu J., Sule L., Li Y., Nucleic Acids Res., 2008, 36(5), 1472—1481 |
| 19 | Sreedhara A., Li Y., Breaker R. R., J. Am. Chem. Soc., 2004,126(11), 3454—3460 |
| 20 | Behera A. K., Schlund K. J., Mason A. J., Alila K. O., Han M., Grout R. L., Baum D. A., Biopolymers, 2013, 99(6), 382—391 |
| 21 | Semlow D. R., Silverman S. K., J. Mol. Evol., 2005, 61(2), 207—215 |
| 22 | Lee Y., Klauser P. C., Brandsen B. M., Zhou C., Li X., Silverman S. K., J. Am. Chem. Soc., 2017, 139(1), 255—261 |
| 23 | Wang M., Zhang H., Zhang W., Zhao Y., Yasmeen A., Zhou L., Yu X., Tang Z., Nucleic Acids Res., 2014, 42(14), 9262—9269 |
| 24 | Camden A. J., Walsh S. M., Suk S. H., Silverman S. K., Biochem., 2016, 55(18), 2671—2676 |
| 25 | Silverman S. K., Acc. Chem. Res., 2015,48(5), 1369—1379 |
| 26 | Walsh S. M., Sachdeva A., Silverman S. K., J. Am. Chem. Soc., 2013, 135(40), 14928—14931 |
| 27 | Walsh S. M., Konecki S. N., Silverman S. K., J. Mol. Evol., 2015, 81(5-6), 218—224 |
| 28 | Chandrasekar J., Wylder A. C., Silverman S. K., J. Am. Chem. Soc., 2015, 137(30), 9575—9578 |
| 29 | Wang P., Silverman S. K., Angew. Chem. Int. Ed., 2016,55(34), 10052—10056 |
| 30 | Chandra M., Sachdeva A., Silverman S. K., Nat. Chem. Biol., 2009, 5(10), 718—720 |
| 31 | Zhou C., Avins J. L., Klauser P. C., Brandsen B. M., Lee Y., Silverman S. K., J. Am. Chem. Soc., 2016,138(7), 2106—2109 |
| 32 | Gu H., Furukawa K., Weinberg Z., Berenson D. F., Breaker R. R., J. Am. Chem. Soc., 2013, 135(24), 9121—9129 |
| 33 | Gu H., Breaker R. R., BioTechniques, 2013, 54(6), 337—343 |
| 34 | Wagenbauer K. F., Sigl C., Dietz H., Nature, 2017, 552(7683), 78—83 |
| 35 | Sednev M. V., Mykhailiuk V., Choudhury P., Halang J., Sloan K. E., Bohnsack M. T., Höbartner C., Angew. Chem. Int. Ed., 2018, 57(46), 15117—15121 |
| 36 | Yu H., Zhang S., Chaput J. C., Nat. Chem., 2012, 4(3), 183—187 |
| 37 | Yan S., Li X., Zhang P., Wang Y., Chen H. Y., Huang S., Yu H., Chem. Sci., 2019,(10), 3110—3117 |
| 38 | Taylor A. I., Holliger P., Nat. Protoc., 2015, 10(10), 1625—1642 |
| 39 | Taylor A. I., Pinheiro V. B., Smola M. J., Morgunov A. S., Peak⁃Chew S., Cozens C., Weeks K. M., Herdewijn P., Holliger P., Nature, 2015, 518(7539), 427—430 |
| 40 | Wang Y., Ngor A. K., Nikoomanzar A., Chaput J. C., Nat. Commun., 2018, 9(1), 5067—5076 |
| 41 | Wang Y., Vorperian A., Shehabat M., Chaput J. C., ChemBioChem, 2020, 21(7), 1001—1006 |
| 42 | Zhou W., Liu J., Metallomics, 2018, 10(1), 30—48 |
| 43 | Lan T., Lu Y., Metal Ions in Life Sciences, 2012,10, 217—248 |
| 44 | Saran R., Chen Q., Liu J., J. Mol. Evol., 2015, 81(5-6), 235—244 |
| 45 | Zhou W., Saran R., Huang P. J., Ding J., Liu J., ChemBioChem, 2017, 18(6), 518—522 |
| 46 | Kasprowicz A., Stokowa⁃Sołtys K., Wrzesiński J., Jeżowska⁃Bojczuk M., Ciesiołka J., Dalton Trans., 2015, 44(17), 8138—8149 |
| 47 | Nelson K. E., Bruesehoff P. J., Lu Y., J. Mol. Evol., 2005, 61(2), 216—225 |
| 48 | Moon W. J., Liu J., ChemBioChem, 2020, 21(3), 401—407 |
| 49 | Liu J., Brown A. K., Meng X., Cropek D. M., Istok J. D., Watson D. B., Lu Y., Proc. Natl. Acad. Sci. USA, 2007, 104(7), 2056—2061 |
| 50 | Zhou W., Zhang Y., Huang P. J., Ding J., Liu J., Nucleic Acids Res., 2016, 44(1), 354—363 |
| 51 | Huang P. J., Vazin M., Liu J., Biochemistry, 2016, 55(17), 2518—2525 |
| 52 | Huang P. J., Vazin M., Matuszek Ż., Liu J., Nucleic Acids Res., 2015, 43(1), 461—469 |
| 53 | Torabi S. F., Wu P., McGhee C. E., Chen L., Hwang K., Zheng N., Cheng J., Lu Y., Proc. Natl. Acad. Sci. USA, 2015, 112(19), 5903—5908 |
| 54 | Zhou W., Saran R., Chen Q., Ding J., Liu J., ChemBioChem, 2016, 17(2), 159—163 |
| 55 | Saran R., Liu J., Anal. Chem., 2016, 88(7), 4014—4020 |
| 56 | Saran R., Kleinke K., Zhou W., Yu T., Liu J., Biochemistry, 2017, 56(14), 1955—1962 |
| 57 | Gu L., Saran R., Yan W., Huang P. J., Wang S., Lyu M., Liu J., ACS Omega, 2018, 3(11), 15174—15181 |
| 58 | Huang P. J., Liu J., Anal. Chem., 2016, 88(6), 3341—3347 |
| 59 | Zhou W., Vazin M., Yu T., Ding J., Liu J., Chemistry, 2016, 22(28), 9835—9840 |
| 60 | Huang P. J., de Rochambeau D., Sleiman H. F., Liu J., Angew. Chem. Int. Ed., 2020, 59(9), 3573—3577 |
| 61 | Wang Y., Liu E., Lam C. H., Perrin D. M., Chem. Sci., 2018, (9), 1813—1821 |
| 62 | Liu M., Chang D., Li Y., Acc. Chem. Res., 2017, 50(9), 2273—2283 |
| 63 | Zhang W., Feng Q., Chang D., Tram K., Li Y., Methods, 2016, 106(15), 66—75 |
| 64 | Devon M., Meghan R., Li Y., Small Methods, 2018, 2(2), 1700319 |
| 65 | Ali M. M., Aguirre S. D., Lazim H., Li Y., Angew. Chem. Int. Ed., 2011, 50(16), 3751—3754 |
| 66 | Shen Z., Wu Z., Chang D., Zhang W., Tram K., Lee C., Kim P., Salena B. J., Li Y., Angew. Chem., Int. Ed., 2016, 55(7), 2431—2434 |
| 67 | Gu L., Yan W., Wu H., Fan S., Ren W., Wang S., Lyu M., Liu J., Anal. Chem., 2019, 91(12), 7887—7893 |
| 68 | He S., Qu L., Shen Z., Tan Y., Zeng M., Liu F., Jiang Y., Li Y., Anal. Chem., 2015, 87(1), 569—577 |
| 69 | Xue P., He S., Mao Y., Qu L., Liu F., Tan C., Jiang Y., Tan Y., Sensors, 2017, 17(3), 650—661 |
| 70 | Geng X., Zhang M., Wang X., Sun J., Zhao X., Zhang L., Wang X., Shen Z., Anal. Chim. Acta, 2020, 1123(1), 28—35 |
| 71 | Chen F.,Bai M.,Cao K., Zhao Y., Wei J., Zhao Y., Adv. Funct. Mater., 2017, 27(45), 1702748 |
| 72 | Chen F.,Bai M.,Cao K.,Zhao Y.,Cao X., Wei J., Wu N., Li J., Wang L., Fan C., Zhao Y., ACS Nano, 2017, 11(12), 11908—11914 |
| 73 | Wang J.,Wang H.,Wang H.,He S.,Li R.,Deng Z., Liu X., Wang F., ACS Nano, 2019, 13(5), 5852—5863 |
| [1] | 张开翔, 刘军杰, 宋巧丽, 王丹钰, 史进进, 张海悦, 李景虹. 基于DNA纳米花的细胞自噬基因沉默用于增敏抗肿瘤化疗[J]. 高等学校化学学报, 2020, 41(7): 1461. |
| [2] | 邬期望, 沈宏. 基于NUPACK预测设计的Toehold诱导链置换反应及其在DNA酶催化微流控化学发光单核苷酸多态性分析中的应用[J]. 高等学校化学学报, 2015, 36(12): 2386. |
| [3] | 封科军, 姚艳玲, 沈国励, 俞汝勤. 基于单壁碳纳米管-DNA酶复合结构的电化学生物传感器[J]. 高等学校化学学报, 2012, 33(08): 1676. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||