

高等学校化学学报 ›› 2021, Vol. 42 ›› Issue (11): 3433.doi: 10.7503/cjcu20210394
收稿日期:2021-06-10
出版日期:2021-11-10
发布日期:2021-11-10
通讯作者:
郑晶
E-mail:zhengjing2013@hnu.edu.cn
基金资助:
CHEN Weiju1, CHEN Shiya2, XUE Caoye3, LIU Bo4, ZHENG Jing2( )
)
Received:2021-06-10
Online:2021-11-10
Published:2021-11-10
Contact:
ZHENG Jing
E-mail:zhengjing2013@hnu.edu.cn
Supported by:摘要:
由于肿瘤内部细胞远离血管, 其氧气消耗量远远超出血液供应量, 因此容易导致肿瘤缺氧. 肿瘤缺氧会引发肿瘤扩散加速、 诱导某些基因过表达及产生药物抗药性等问题. 基于此, 发展性能优异的缺氧响应荧光探针对肿瘤的诊断和治疗具有重要意义. 本文对缺氧响应荧光探针在成像及治疗方面的应用进展进行了综合评述, 介绍了硝基、 偶氮键和醌3种常用的缺氧响应基团, 并探讨了它们在缺氧微环境下的识别机理; 介绍了缺氧响应荧光探针的构建及其在生物成像方面的最新研究成果; 总结了缺氧响应荧光探针在基因治疗、 光动力学治疗、 化学治疗及协同治疗方面的研究进展; 展望了缺氧响应荧光探针在临床诊断和治疗方面的应用前景.
中图分类号:
TrendMD:
谌委菊, 陈诗雅, 薛曹叶, 刘波, 郑晶. 缺氧响应荧光探针的成像及治疗应用. 高等学校化学学报, 2021, 42(11): 3433.
CHEN Weiju, CHEN Shiya, XUE Caoye, LIU Bo, ZHENG Jing. Fluorescent Probe for Hypoxia-triggered Imaging and Cancer Therapy. Chem. J. Chinese Universities, 2021, 42(11): 3433.
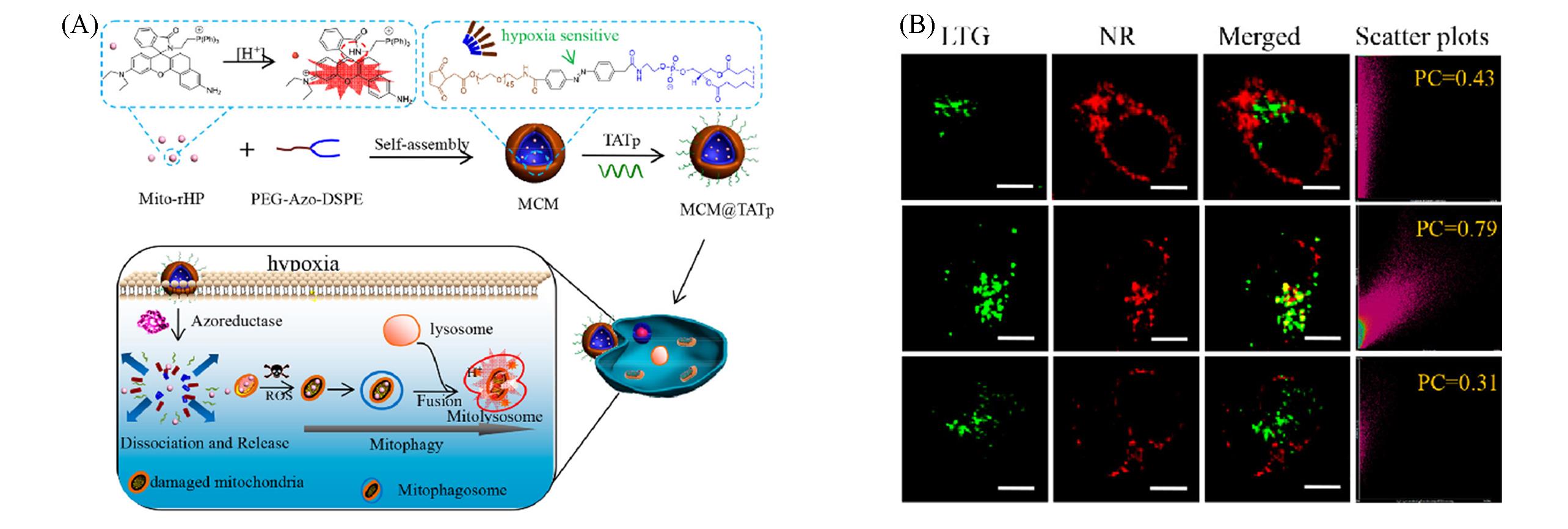
Fig.7 Schematic illustration of azo?based amphiphilic polymer and its work principle in hypoxia cell(A) and confocal fluorescence microscopy imaging of cells incubated with polymer under different treatments(B)[46]Copyright 2019, American Chemical Society.
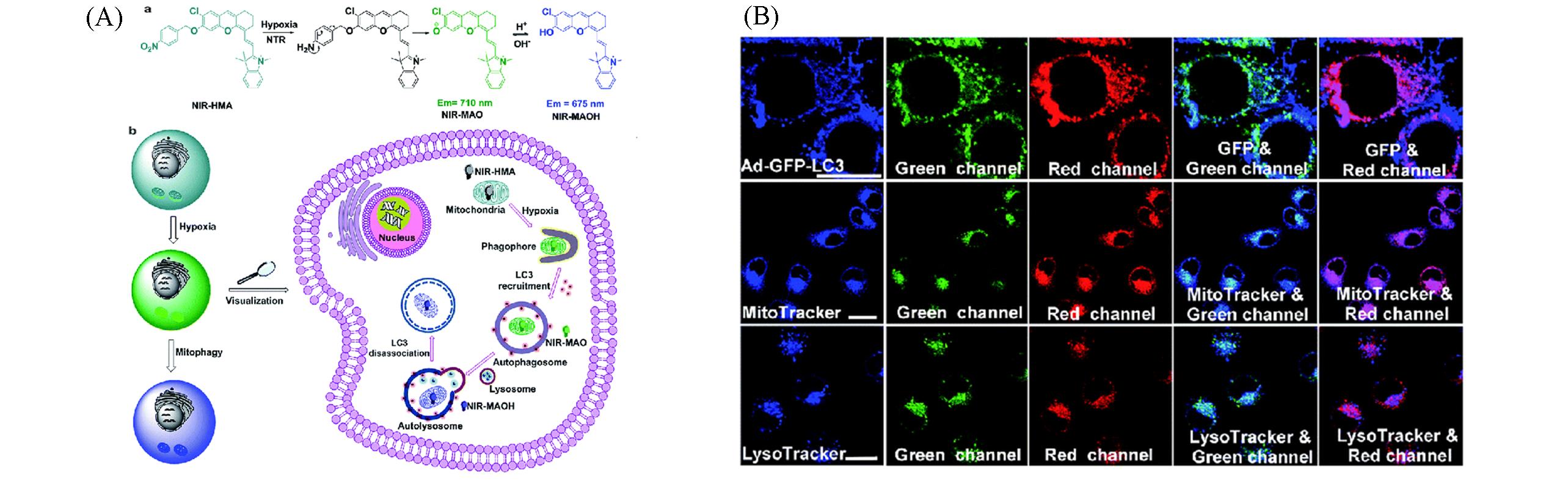
Fig.8 Schematic diagram of NIR probe for visualization of hypoxia induced mitophagy(A) and fluorescence images for tracking hypoxia induced mitophagy process(B)[47]Copyright 2018, Royal Society of Chemistry.
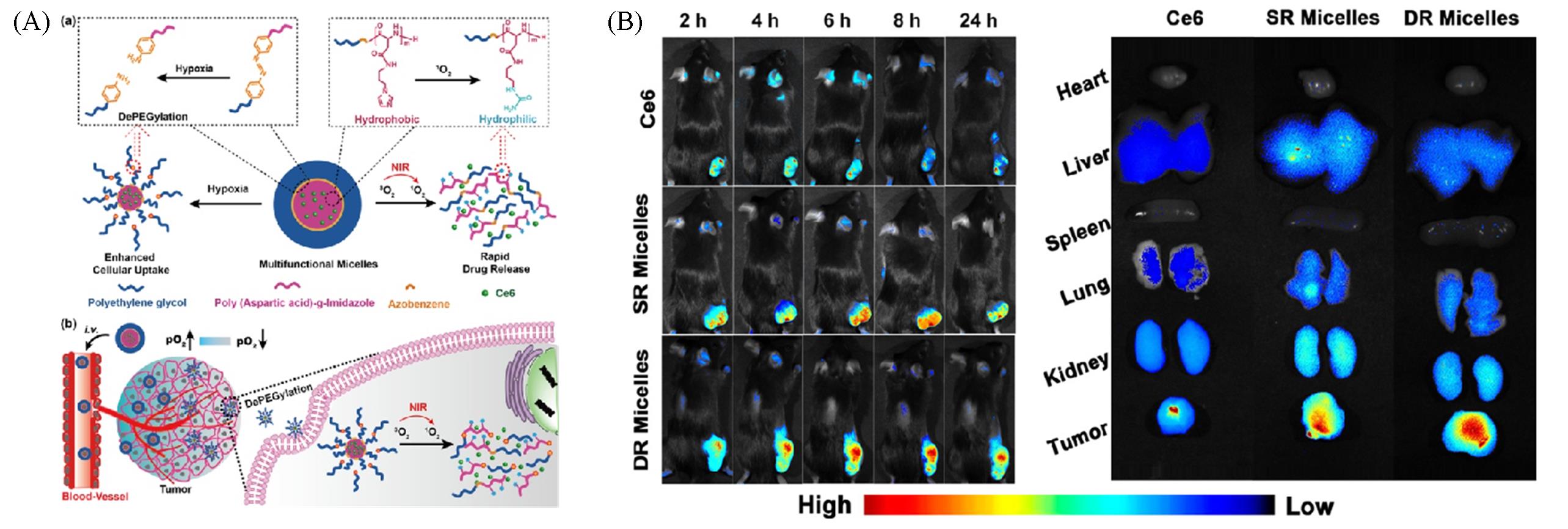
Fig.10 Illustration of hypoxia?sensitive micelles for photodynamic therapy(A) and fluorescent imaging of tumor organs and healthy organs under different treatment(B)[56]Copyright 2018, American Chemical Society.
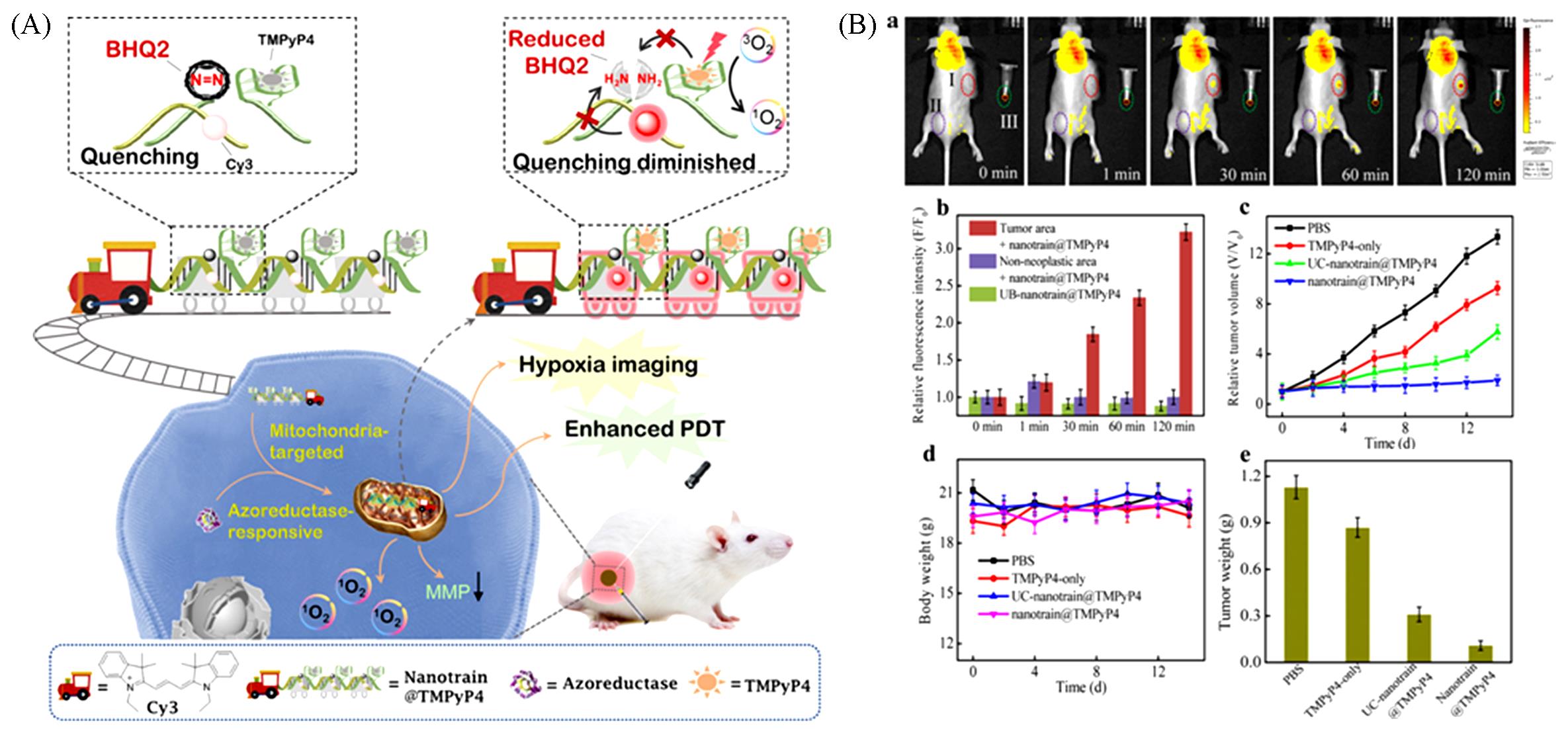
Fig.11 Schematic illustration of the azoreductase?responsive, mitochondrion?targeted probe for hypoxia imaging and enhanced PDT(A) and the therapy efficacy of the mitochondrion?targeted probe in representative mice(B)[57]Copyright 2021, American Chemical Society.
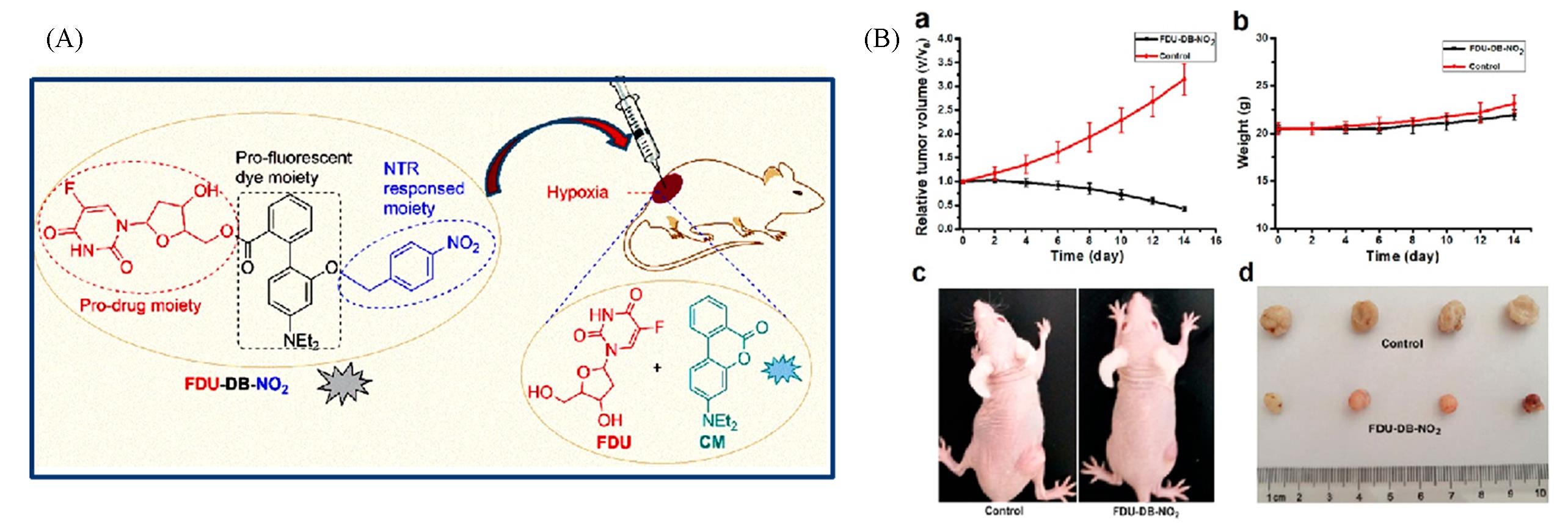
Fig.12 Schematic illustration for triggered release of hypoxia?responsive drug delivery system(A) and the therapy efficacy of the drug delivery system after different treatment(B)[64]Copyright 2018, American Chemical Society.
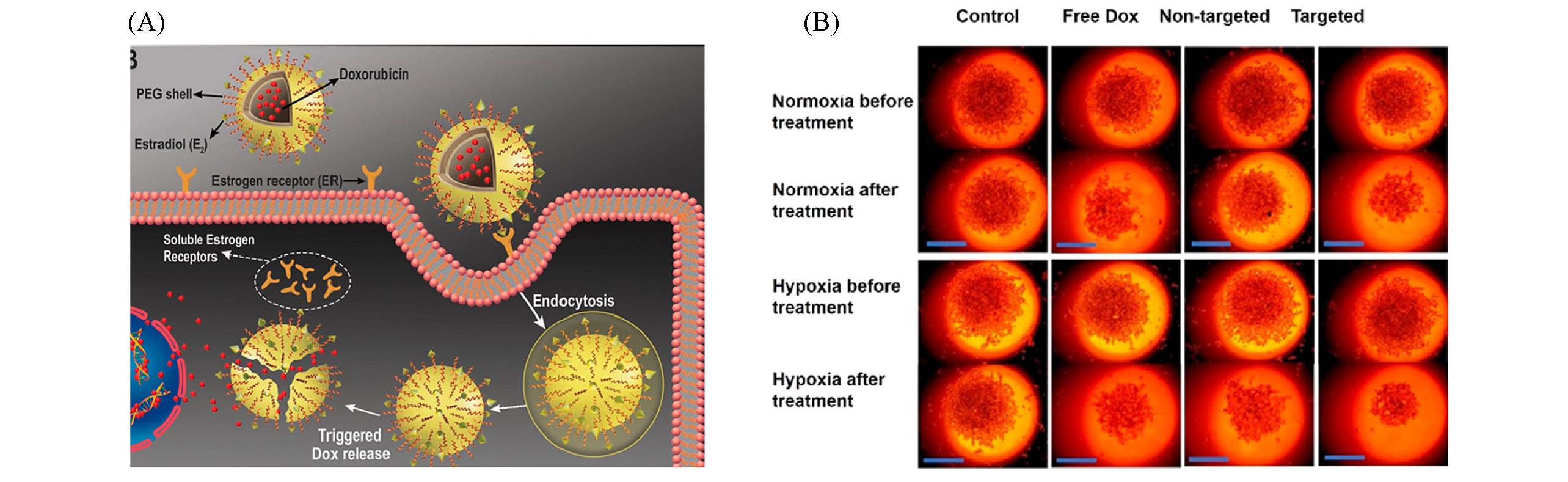
Fig.13 Schematic illustration of the targeted DOX?oaded polymersomes for chemotherapy(A) and representative images of the MCF7 cells with different treatment(B)[65]Copyright 2020, American Chemical Society.
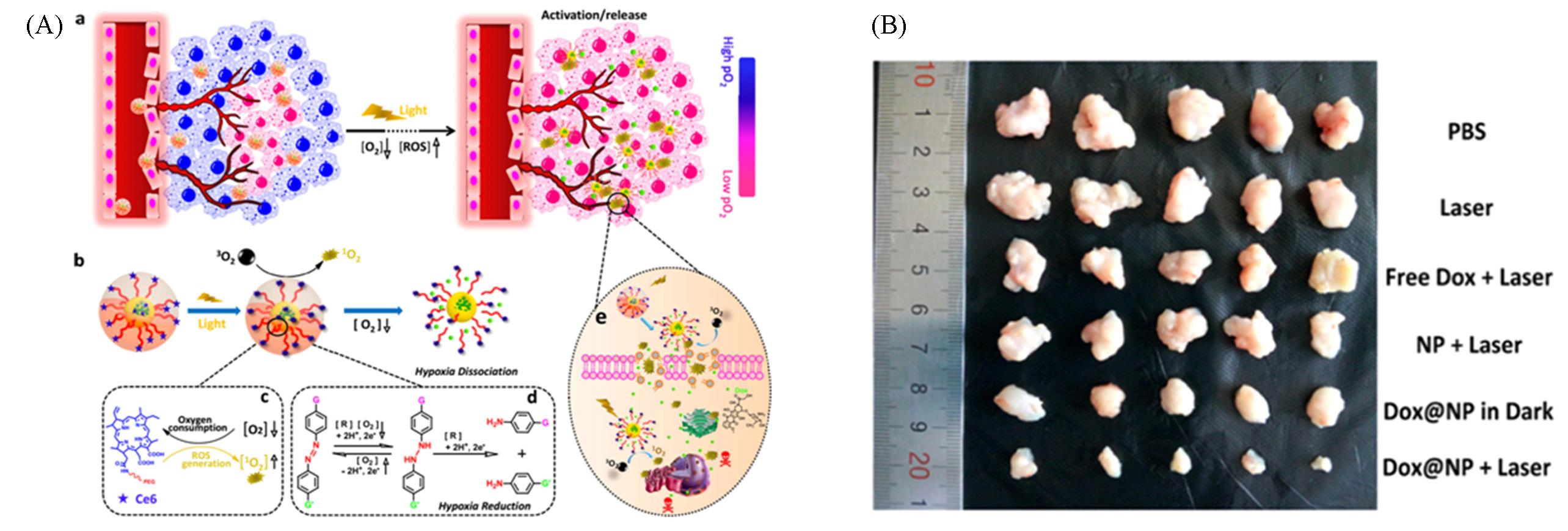
Fig.14 Schematic of the hypoxia?triggered DOX@NP nanoparticles for synergistic therapy treatment(A) and representative images of the tumor after different treatments(B)[66]Copyright 2018, American Chemical Society.
| Sensing moiety | Trigger | Reduction mechanism | Application | Ref. |
|---|---|---|---|---|
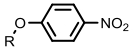 | Nitroreductase | 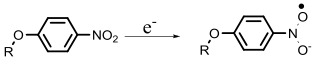 | Hypoxia?related parameter imaging, metabolic process imaging | [20, 24, 36—39, 47] |
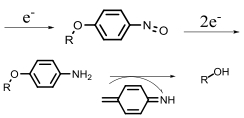 | Chemotherapy, photodynamic therapy | [ | ||
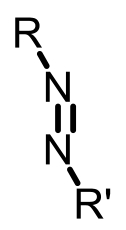 | Azoreductase | 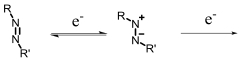 | Hypoxia?related parameter imaging,metabolic process imaging | [ |
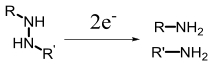 | Gene therapy, chemotherapy, photodynamic therapy, synergistic therapy | [29, 50, 55—57, 63, 65—69] | ||
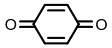 | Quinone reductase | 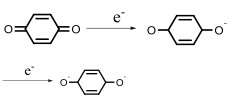 | Hypoxia?related parameter imaging, metabolic process imaging | [ |
Table 1 Summary of typical hypoxia-triggered probes and their applications
| Sensing moiety | Trigger | Reduction mechanism | Application | Ref. |
|---|---|---|---|---|
 | Nitroreductase | 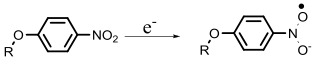 | Hypoxia?related parameter imaging, metabolic process imaging | [20, 24, 36—39, 47] |
 | Chemotherapy, photodynamic therapy | [ | ||
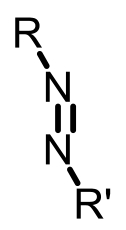 | Azoreductase |  | Hypoxia?related parameter imaging,metabolic process imaging | [ |
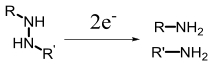 | Gene therapy, chemotherapy, photodynamic therapy, synergistic therapy | [29, 50, 55—57, 63, 65—69] | ||
 | Quinone reductase |  | Hypoxia?related parameter imaging, metabolic process imaging | [ |
| 49 | Williamson B., Nature, 1982, 298(5873), 416—418 |
| 50 | Perche F., Biswas S., Wang T., Zhu L., Torchilin V. P., Angew. Chem. Int. Ed., 2014, 53(13), 3362—3366 |
| 51 | Li X., Kwon N., Guo T., Liu T., Yoon J., Chem. Mater., 2018, 57(36), 11522—11531 |
| 52 | Md N., Chung S. J., Kalashnikova I., Meghan L. H., Silver H., George J., Christopher H. C., Kim T., ACS Applied. Bio. Materials, 2020, 3(11), 7989—7999 |
| 53 | Liu Y. Y., Liu Y., Bu W. B., Cheng C., Zuo C. J., Xiao Q. F., Sun Y., Ni D. L., Zhang C., Liu J. N., Shi J. L., Angew. Chem. Int. Ed., 2015, 54(28), 8105—8109 |
| 54 | Li J., Ou H.L., Dan D., Chem. Res. Chinese Universities, 2021, 37(1), 83—89 |
| 55 | Li J., Liu W., Li Z. H., Hu Y. C., Yang J. F., Li J. S., Chem. Commun.,2021, 57(38), 4710—4713 |
| 56 | Li J. J, Meng X., Jian D., Lu D., Zhang X., Chen Y. R, Zhu J. D., Fan A. P., Ding D., Kong D. L., Wang Z., Zhao Y. J., ACS Appl. Mater. Interfaces, 2018, 10(20), 17117—17128 |
| 57 | Liu J., Ding G., Chen S. Y., Xue C. Y., Chen M., Wu X., Yuan Q., Zheng J., Yang R. H., ACS Appl. Mater. Interfaces, 2021, 13(8), 9681—9690 |
| 58 | Chen H. C., Bi Q. R., Yao Y. R., Tan N. H., J. Mater. Chem. B, 2018, 6(26),4351—4359 |
| 59 | Sui M. H., Liu W. W., Shen Y. Q., J. Control. Release, 2011, 155(2) 227—236 |
| 60 | Hu Q. Y., Sun W. J., Wang C., Gu Z., Adv. Drug Delivery Rev., 2016, 98, 19—34 |
| 61 | Li M. Q., Ma H., Shi C., Zhang H., Long S., Sun W., Du J. J., Fan J. L., Peng X. J., Chem. Res. Chinese Universities, 2021, 37(4),doi: 10.1007/s40242⁃021⁃1186⁃3 |
| 62 | Li H. L., Lei W. Y., Wu J. N., Li S. H., Zhou G. Q., Liu D. D., Yang X. J., Wang S. X., Li Z. H., Zhang J. C., J. Mater. Chem. B, 2018, 6(18), 2747—2757 |
| 63 | Lee J., Oh E. T., Yoon H., Kim C. W., Han Y., Song J., Jang H., Park H. J., Kim C., Nanoscale, 2017, 9(20), 6901—6909 |
| 64 | Liu W., Liu H. T., Peng X. R., Zhou G. Q., Liu D. D., Li S. H., Zhang J. C., Wang S. X., Bioconjugate. Chem., 2018, 29(10), 3332—3343 |
| 65 | Mamnoon B., Feng L., Froberg J., Choi Y., Sathish V., Mallik S., Mol. Pharm., 2020, 17(11), 4312—4322 |
| 1 | Brown J. M., Wilson W. R., Nat. Rev. Cancer, 2004, 4(6), 437—447 |
| 2 | Harris A. L., Nat. Rev. Cancer, 2002, 2(1), 38—47 |
| 3 | Lee P., Chandel N. S., Simon M. C., Nat. Rev. Mol. Cell. Biol., 2020, 21(5), 268—283 |
| 4 | Yun J. K., McCormic T. S., Villabona C., Judware R. R., Espinosa M. B., Lapetina E. G., Proc. Natl. Acad. Sci., USA, 1997, 94(25),13903—13908 |
| 5 | Mazure N. M., Pouyssegur J., Curr. Opin. Cell Biol., 2010, 22(2), 177—180 |
| 6 | Hu Y. L., Delay M., Jahangiri A., Molinaro A. M., Rose S. D., Carbonell W. S., Aghi M. K., Cancer Res., 2012, 72(7), 1773—1783 |
| 7 | Wilson W. R., Hay M. P., Nat. Rev. Cancer, 2011, 11(6), 393—410 |
| 8 | Sahu A., Kwon I., Tae G., Biomaterials,2020, 228, 119578 |
| 9 | Liu Y. Y., Jiang Y. Q., Zhang M., Tang Z. M., He M. Y., Bu W., Acc. Chem. Res., 2018, 51(10), 2502—2511 |
| 10 | Zbaida S., Levin W. G., Chem. Res. Toxicol., 1991, 4(1), 82—88 |
| 11 | Boddu R. S., Perumal O., K D., Biotechnol. Appl. Biochem., 2020, doi: 10.1002/bab.2073 |
| 12 | Yang Y., Zhang Y., Wu Q. Y., Cui X. L., Lin, Z. H., Liu S. P., Chen L. Y., J. Exp. Clin. Cancer Res., 2014, 33, 14 |
| 13 | Thambi T., Park J. H., Lee D. S., Chem. Commun., 2016, 52(55), 8492—8500 |
| 66 | Wang W. L., Lin L., Ma X. J., Wang B., Liu S. R., Yan X. X., Li S. R., Tian H. Y., Yu X. F., Appl. Mater. Interfaces, 2018, 10(23), 19398—19407 |
| 67 | Zhang X. L., Wu M., Li J., Lan S. Y., Zeng Y. Y., Liu X. L., Liu J. F., Appl. Mater. Interfaces,2018, 10(26),21909—21919 |
| 68 | Huang C. X., Zheng J., Ma D. D., Liu N., Zhu C., Li J. S., Yang R. H., J. Mater. Chem. B, 2018, 6(40), 6424—6430 |
| 14 | Sanduleanu S., Wiel A., Lieverse R. I. Y., Marcus D., Ibrahim A., Primakov S., Wu G. Y., TheysJ., Yaromina A., Dubois L. J., Lambin P., Cancers, 2020, 12(5), 1322 |
| 15 | Bennani⁃Baiti B., Pinker K., Zimmermann M., Helbich T. H., Baltzer P. A., Clauser P., Kapetas P., Bago⁃Horvath Z., Stadlbauer A., Cancers, 2020, 12(8), 2024 |
| 16 | Sun X. L., Niu G., Chan N., Shen B. Z., Chen X. Y., Mol. Imaging Biol., 2011, 13(3), 399—410 |
| 17 | Gao P., Pan W., Li N., Tang B., Chem. Sci., 2019, 10(24), 6035—6071 |
| 18 | Zhang J., Liu H. W., Hu X. X., Li J., Liang L. H., Zhang X. B., Tan W. H., Anal. Chem., 2015, 87(23), 11832—11839 |
| 69 | Huang C. X., Tan W. H., Zheng J., Zhu C., Huo J., Yang R. H., Appl. Mater. Interfaces, 2019, 11( 29), 25740—25749 |
| 70 | Riordan J. R., Deuchars K., Kartner N., Alon N., Trent J., Ling V., Nature, 1985, 316(6031), 817—819 |
| 19 | Munkholm C., Parkinson D. R., Walt D. R., J. Am. Chem. Soc.,1990, 112( 7), 2608—2612 |
| 20 | Li Z., Li X. H., Gao X. H., Zhang Y. Y., Shi W., Ma H. M., Anal. Chem., 2013, 85(8), 3926—3932 |
| 71 | Shukla S. K., Purohit V., Mehla K., Gunda V., Chaika N. V., Vernucci E., King R. J., Abrego J., Goode G. D., Dasgupta A., Illies A. L., Gebregiworgis T., Dai B. B., Augustine J. J., Murthy D., Attri K. S., Mashadova O., Grandgenett P. M., Powers R., Ly Q. P., Lazenby A. J., Grem J. L., Yu F., Mates J. M., Asara J. M., Kim J. W., Hankins J. H., Weekes C., Hollingsworth M. A., Serkova N. J., Sasson A. R., Fleming J. B., Oliveto J. M., Lyssiotis C. A., Cantley L. C., Berim L., Singh P. K., Cancer Cell, 2017,32(3), 392 |
| 72 | Tian H., Luo Z. Y., Liu L. L., Zheng M. B., Chen Z., Ma A. Q., Liang R. J., Han Z. Q., Lu C. Y., Cai L. T., Adv. Funct. Mater., 2017, 27, 1703197 |
| 21 | Li Y. H., Wang Y. J., Yang S., Zhao Y. R., Yuan L., Zheng J., Yang R. H., Anal. Chem., 2015, 87(4), 2495—2503 |
| 22 | He X. Y., Li L. H., Fang Y., Shi W., Li X. H., Ma H. M., Chem. Sci., 2017, 5( 8), 3479—3783 |
| 23 | Yu X. Z, Xiang L., Yang S., Qu S.L., Zeng X., Zhou Y. B., Yang R. H., Spectrochim. Acta, Part A, 2021, 245, 118887 |
| 24 | Fan Y. S., Lu M., Yu X. A., He M. L., Zhang Y., Ma X. N., Kou J. P., Yu B.Y., Tian J. W., Anal. Chem., 2019, 91(10), 6585—6592 |
| 25 | Eom T., Yoo W., Kim S., Khan A., Biomaterials,2018, 185, 333—347 |
| 26 | Rao J., Khan A., J. Am. Chem. Soc., 2013, 135(38), 14056—14059 |
| 27 | Kiyose K., Hanaoka K., Oushiki D., Nakamura T., Kajimura M., Suematsu M., Nishimatsu H., Yamane T., Terai T., Hirata Y., Nagano T., J. Am. Chem. Soc., 2010, 132(45), 15846—15848 |
| 28 | Tian Y., Li, Y. F., Jiang W. L., Zhou D. Y., Fei J. J., Li C. Y., Anal. Chem.,2019, 91(16), 10901—10907 |
| 29 | Zhao X. B., Ha W., Gao K., Shi Y. P., Anal. Chem., 2020, 92(13), 9039—9047 |
| 30 | Piao W., Tsuda S., Tanaka Y., Maeda S., Liu F. Y., Takahashi S., Kushida Y., Komatsu T., Ueno T., Terai T., Angew. Chem. Int. Ed., 2013, 52(49), 13028—13032 |
| 31 | Wendlandt A. E., Stahl S. S., Angew. Chem. Int. Ed.,2015, 54(49), 14638—14658 |
| 32 | Singh S. K., Husain S. M., ChemBioChem, 2018, 19(15), 1657—1663 |
| 33 | Sun L. L., Chen Y., Kuang S., Li G.Y., Guan R.L., Liu J. P., Ji L. N., Chao H., Chem. Eur. J., 2016, 22, 8955—8965 |
| 34 | Komatsu H., Harada H., Tanabe K., Hiraoka M., Nishimoto S., Med. Chem. Commun., 2010, 1, 50 Ch |
| 35 | Zhang P. Y., Huang H. Y., Chen Y., Wang J. Q., Ji L. N., Chao H., Biomaterials, 2015, 53, 522teri |
| 36 | Zheng A. X., Sun H. Y., Du Y. L., Wang Y. R., Wu M., Liu X. L., Zeng Y. Y., Liu J. F., Anal. Chim. Acta,2021, 1144, 76—84 |
| 37 | Fan L., Zan Q., Lin B., Wang X. D., Gong X. J., Zhao Z. H., Shuang S. M., Dong C., Wong M .S., Analyst, 2020, 145(16), 5657—5663 |
| 38 | Wang Y., Han X. Y., Zhang X., Zhang L., Chen L. X., Analyst, 2020, 145(4), 1389—1395 |
| 39 | Zhang X. Q., Zhao Q., Li Y. R., Duan X. R., Tang Y. L., Anal. Chem.,2017, 89(10), 5503—5510 |
| 40 | Lan G. X., Ni K. Y., You E., Wang M. L., Culbert A., Jiang X. M., Lin W. B., J. Am. Chem. Soc., 2019, 141(48), 18964—18969 |
| 41 | Pouyssegur J., Dayan F., Mazure N. M., Nature, 2006, 441(7092), 437—443 |
| 42 | Sim J., Cowburn A. S., Palazon A., Madhu B., Tyrakis P. A., Macias D., Bargiela D. M., Pietsch S., Gralla M., Evans C. E., Kittipassorn T., Chey Y. C. J., Branco C. M., Rundqvist H., Peet D. J., Johnson R. S., Cell Metab., 2018, 27(4), 898—913 |
| 43 | Liu T., Laurell C., Selivanova G., Lundeberg J., Nilsson P., Wiman K. G., Cell Death Differ., 2007, 14(3), 411—421 |
| 44 | Yadav A. K., Yadav P. K., Chaudhary G. R., Tiwari M., Gupta A., Sharma A., Pandey A. N., Pandey A. K., Chaube S. K., Cell Mol. Life Sci., 2019, 76(17), 3311—3322 |
| 45 | Li X. Y., Li Y. H., Ma H. M., Chem. Sci., 2020, 11(6), 1617—1622 |
| 46 | Ma D. D., Huang C. X., Zheng J., Zhou W. Y., Tang J. R., Chen W. J., Li J. S., Yang R. H., Anal. Chem., 2019, 91(2), 1360—1367 |
| 47 | Liu Y. C., Teng L. L., Chen L. L., Ma H. C., Liu H. W., Zhang X. B., Chem. Sci., 2018,9(24), 5347—5353 |
| 48 | Chen S. Y., Liu J., Li Y. H., Wu X., Yuan Q., Yang R. H., Zheng J., Trac⁃trend. Anal. Chem., 2020, 131, 116010 |
| [1] | 赵永梅, 穆叶舒, 洪琛, 罗稳, 田智勇. 双萘酰亚胺衍生物用于检测水溶液中的苦味酸[J]. 高等学校化学学报, 2022, 43(3): 20210765. |
| [2] | 唐倩, 但飞君, 郭涛, 兰海闯. 喹啉酮-香豆素类Hg2+比色荧光探针的合成及应用[J]. 高等学校化学学报, 2022, 43(2): 20210660. |
| [3] | 王迪, 钟克利, 汤立军, 侯淑华, 吕春欣. 席夫碱共价有机框架的合成及对I ‒ 的识别[J]. 高等学校化学学报, 2022, 43(10): 20220115. |
| [4] | 黄珊, 姚建东, 宁淦, 肖琦, 刘义. 石墨烯量子点荧光探针对碱性磷酸酶活性的高效检测[J]. 高等学校化学学报, 2021, 42(8): 2412. |
| [5] | 李安然, 赵冰, 阚伟, 宋天舒, 孔祥东, 卜凡强, 孙立, 殷广明, 王丽艳. 基于菲并咪唑的ON⁃OFF⁃ON双比色荧光探针及细胞成像[J]. 高等学校化学学报, 2021, 42(8): 2403. |
| [6] | 杨新杰, 赖艳琼, 李秋旸, 张艳丽, 王红斌, 庞鹏飞, 杨文荣. 基于环状DNA-银纳米簇荧光探针对微囊藻毒素-LR的传感检测[J]. 高等学校化学学报, 2021, 42(12): 3600. |
| [7] | 黄加玲,刘凤娇,王婷婷,刘翠娥,郑凤英,王振红,李顺兴. 氮硫共掺杂碳量子点对胃液pH值的精确检测[J]. 高等学校化学学报, 2020, 41(7): 1513. |
| [8] | 袁中文, 贺利贞, 陈填烽. 单原子催化剂的生物医学应用[J]. 高等学校化学学报, 2020, 41(12): 2690. |
| [9] | 郑有为, 田菲, 张倩, 徐迪, 杨国海, 渠陆陆. 基于SERS纳米探针的细胞内硝基还原酶检测[J]. 高等学校化学学报, 2020, 41(12): 2742. |
| [10] | 吴倩, 程丹, 吕芸, 袁林, 张晓兵. 大斯托克斯位移远红光至近红外荧光探针用于检测肝损伤过程中过氧化亚硝酸盐的动态变化[J]. 高等学校化学学报, 2020, 41(11): 2426. |
| [11] | 王金金, 戚少龙, 杜建时, 杨清彪, 宋岩, 李耀先. 苯并噻唑类荧光探针的合成及对N2H4·H2O和HS |
| [12] | 张勇,申城,幸志荣,陈归柒,卢资,侯志兵,陈雪梅. 可视化检测次氯酸的苯并咪唑类荧光增强型探针[J]. 高等学校化学学报, 2019, 40(12): 2480. |
| [13] | 张莹莹,黄译文,赵冰,王丽艳,宋波. Cr 3+比色荧光探针的合成及细胞成像应用[J]. 高等学校化学学报, 2019, 40(12): 2486. |
| [14] | 喻照川, 马文辉, 吴涛, 问婧, 张永, 王丽艳, 初红涛. B,N,S共掺杂石墨烯量子点的制备及对Fe 3+和H2P |
| [15] | 刘大海, 张雪妍, 冯玉沙, 杜显龙, 薛龙启, 杜建时, 张桂荣, 杨清彪, 李耀先. 基于荧光素硫代酰肼类Hg2+荧光探针的合成及在生物成像中的应用[J]. 高等学校化学学报, 2018, 39(7): 1412. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||