

高等学校化学学报 ›› 2022, Vol. 43 ›› Issue (12): 20220572.doi: 10.7503/cjcu20220572
赵雪琪1,2, 赵越2, 薛静2, 白敏2, 陈锋2, 孙颖1, 宋大千1( ), 赵永席2(
), 赵永席2( )
)
收稿日期:2022-08-29
出版日期:2022-12-10
发布日期:2022-10-25
通讯作者:
宋大千,赵永席
E-mail:songdq@jlu.edu.cn;yxzhao@mail.xjtu.edu.cn
基金资助:
ZHAO Xueqi1,2, ZHAO Yue2, XUE Jing2, BAI Min2, CHEN Feng2, SUN Ying1, SONG Daqian1( ), ZHAO Yongxi2(
), ZHAO Yongxi2( )
)
Received:2022-08-29
Online:2022-12-10
Published:2022-10-25
Contact:
SONG Daqian, ZHAO Yongxi
E-mail:songdq@jlu.edu.cn;yxzhao@mail.xjtu.edu.cn
Supported by:摘要:
单细胞成像可在单细胞水平观测目标物位置、 确定目标物含量, 在生命科学与临床医学研究领域应用广泛. 核酸编码扩增技术利用特定分子反应将待测目标识别转化为核酸条码的扩增, 具有探针种类多、 易编程、 反应条件温和及信号放大效率高等特点, 在单细胞低丰度、 高灵敏、 多目标物成像中优势显著, 为理解细胞状态、 探索生命过程提供了新思路. 本文综合评述了核酸编码扩增在单细胞荧光成像领域的研究进展, 以目标物的编码方式为分类依据, 系统阐述了固定细胞原位成像和活细胞成像中不同目标物编码与扩增成像方式的区别, 并对活细胞成像中多重检测面临的问题以及未来发展前景进行了展望.
中图分类号:
TrendMD:
赵雪琪, 赵越, 薛静, 白敏, 陈锋, 孙颖, 宋大千, 赵永席. 单细胞核酸编码扩增成像分析. 高等学校化学学报, 2022, 43(12): 20220572.
ZHAO Xueqi, ZHAO Yue, XUE Jing, BAI Min, CHEN Feng, SUN Ying, SONG Daqian, ZHAO Yongxi. Nucleic Acids-encoded Amplification for Single-cell Imaging. Chem. J. Chinese Universities, 2022, 43(12): 20220572.
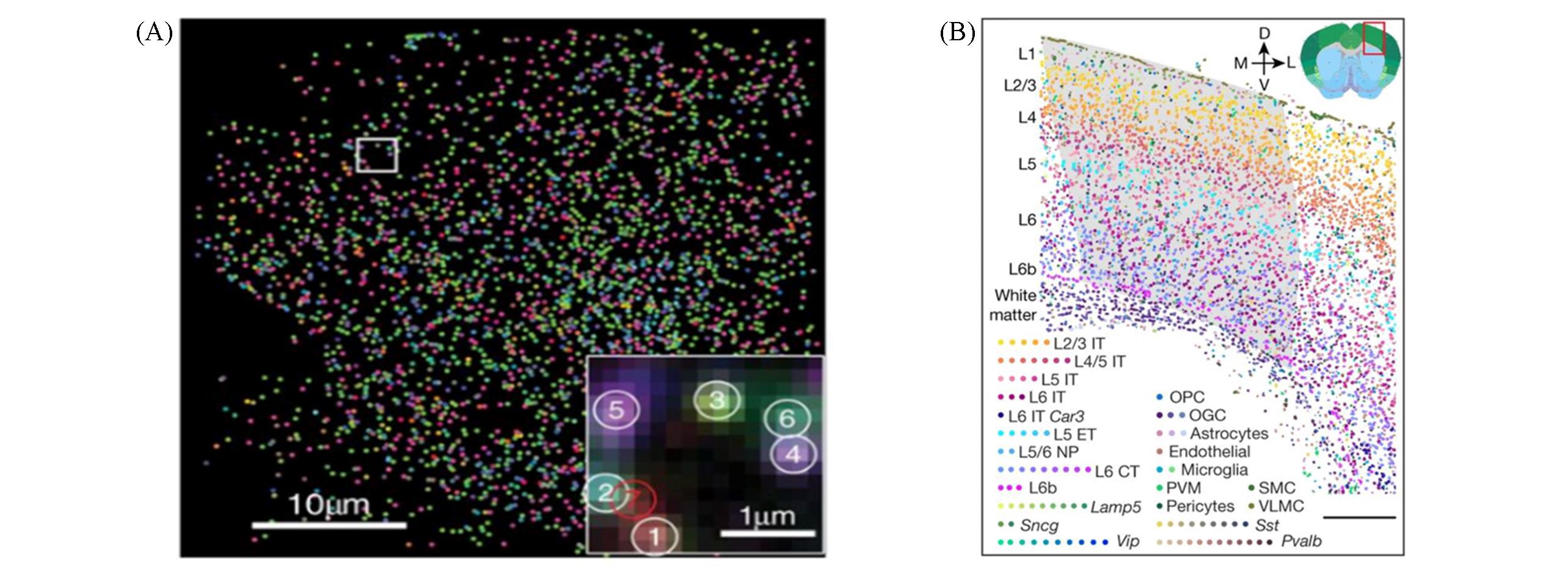
Fig.3 Data of application of MERFISH(A) The composite, false-colored fluorescent image of all detected single molecules in this cell according to their measured binary words[37]; (B) spatial map of the cell clusters in a coronal slice[40].(A) Copyright 2015, the American Association for the Advancement of Science; (B) Copyright 2021, Springer Nature.
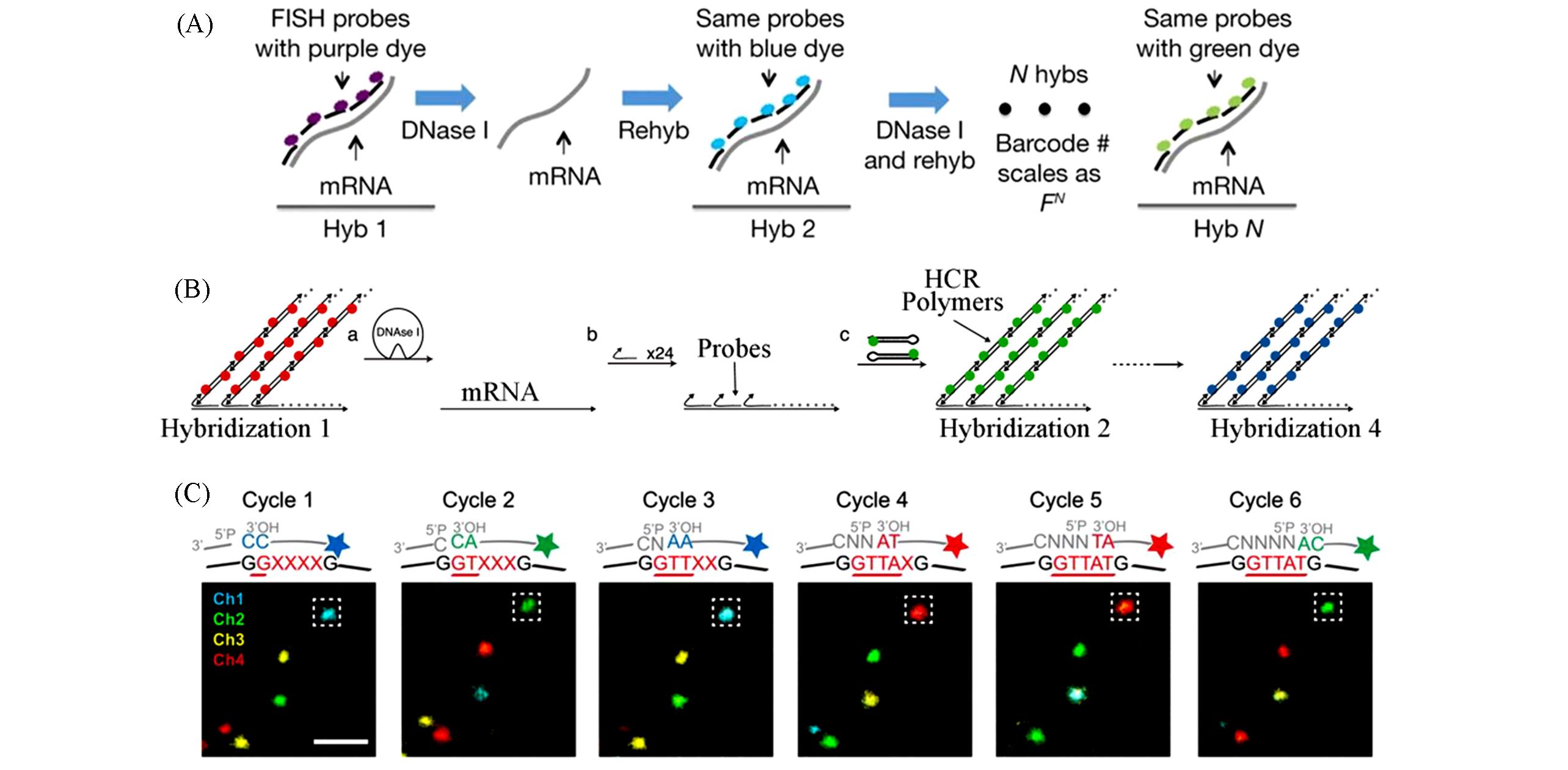
Fig.4 Schematic of hybridization encoded amplification methods(A) Schematic of seqFISH[42]; (B) schematic of HCR-seqFISH[43]; (C) schematic of STARmap[44].(A) Copyright 2014, Springer Nature; (B) Copyright 2016, Elsevier; (C) Copyright 2018, the American Association for the Advancement of Science.
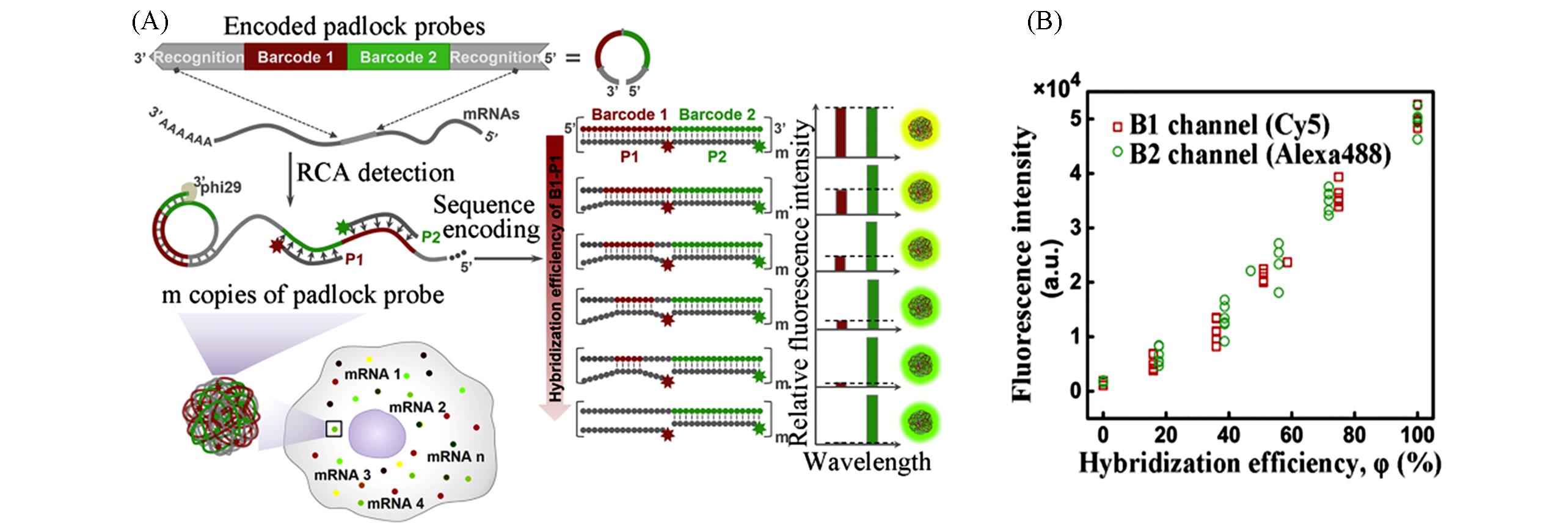
Fig.5 Schematic and data of SeqEA[46](A) Schematic of SeqEA for multiplexed imaging of single-molecule RNAs in single cells; (B) the relationship between hybridization efficiency and fluorescence intensity.Copyright 2018, Elsevier.
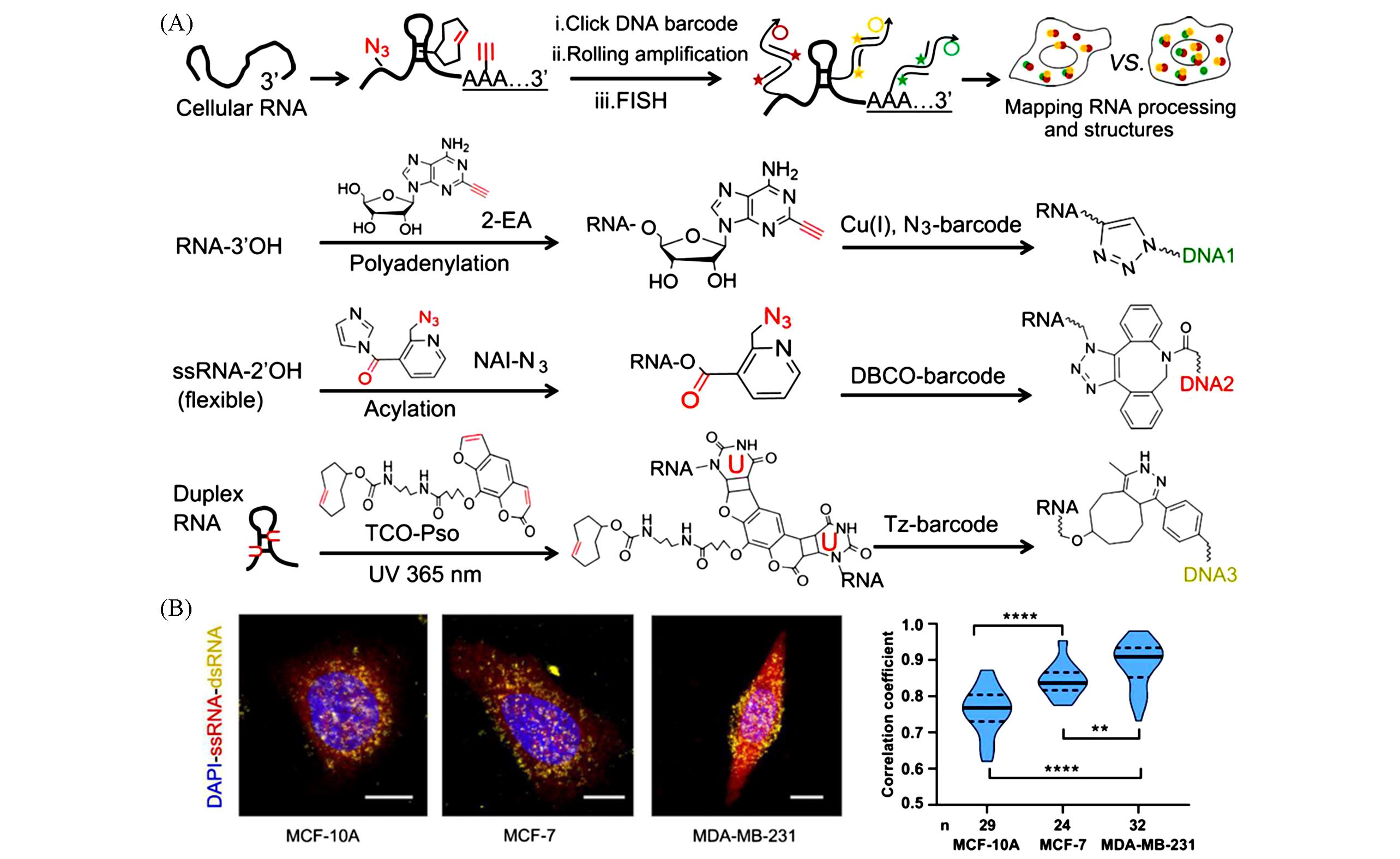
Fig.6 Schematic and data of Clicker⁃FISH[53](A) Schematic of Clicker-FISH; (B) representative cell images from different cell typed and the violin plot for correlation coefficients between ssRNA and dsRNA in these cell types. Copyright 2019, the authors.
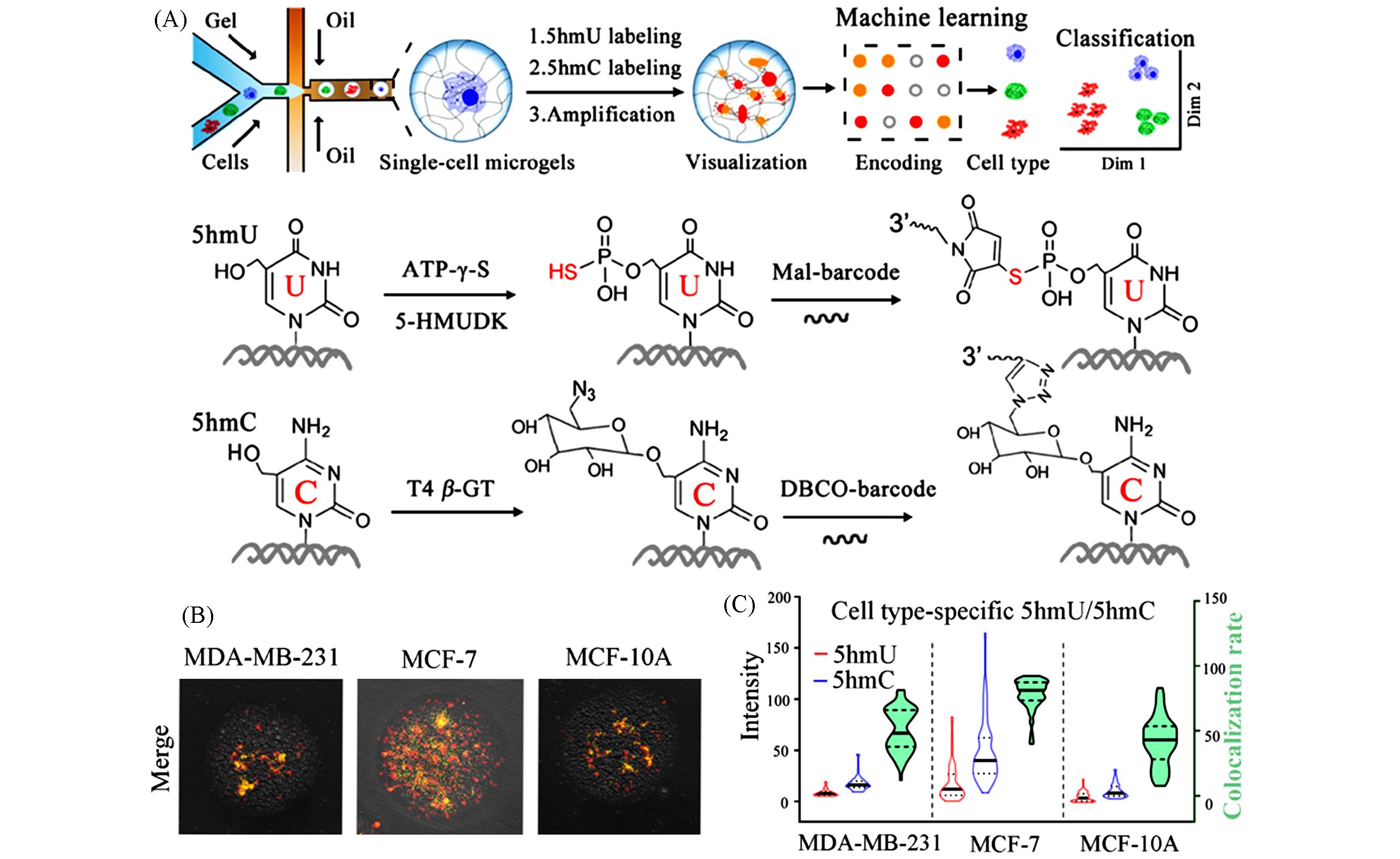
Fig.7 Schematic and data of sc5hmU/5hmC⁃microgel[23](A) Schematic of differentiated visualization of 5hmU and 5hmC with microgel encoding in single cells; (B) representative single-cell microgel images; (C) the intensity and colocalization analysis for 5hmU and 5hmC in different cell lines.Copyright 2020, American Chemical Society.
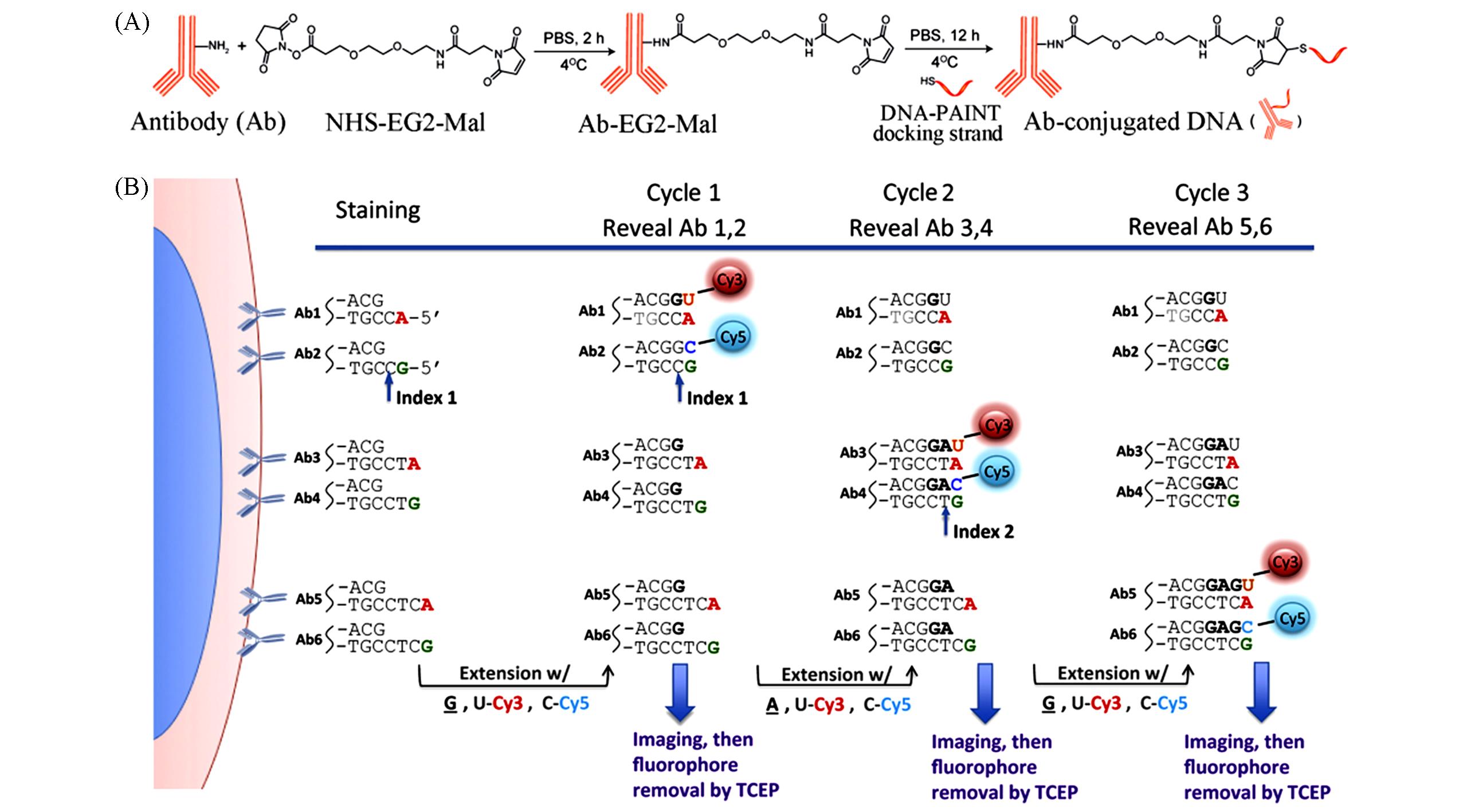
Fig.8 Schematic of antigen⁃antibody encoding methods(A) Workflow of barcodes modified on antibodies for DNA-PAINT[56]; (B) schematic of CODEX(co-detection by indexing) multiplexed imaging[58].(A) Copyright 2017, the authors; (B) Copyright 2018, the authors.
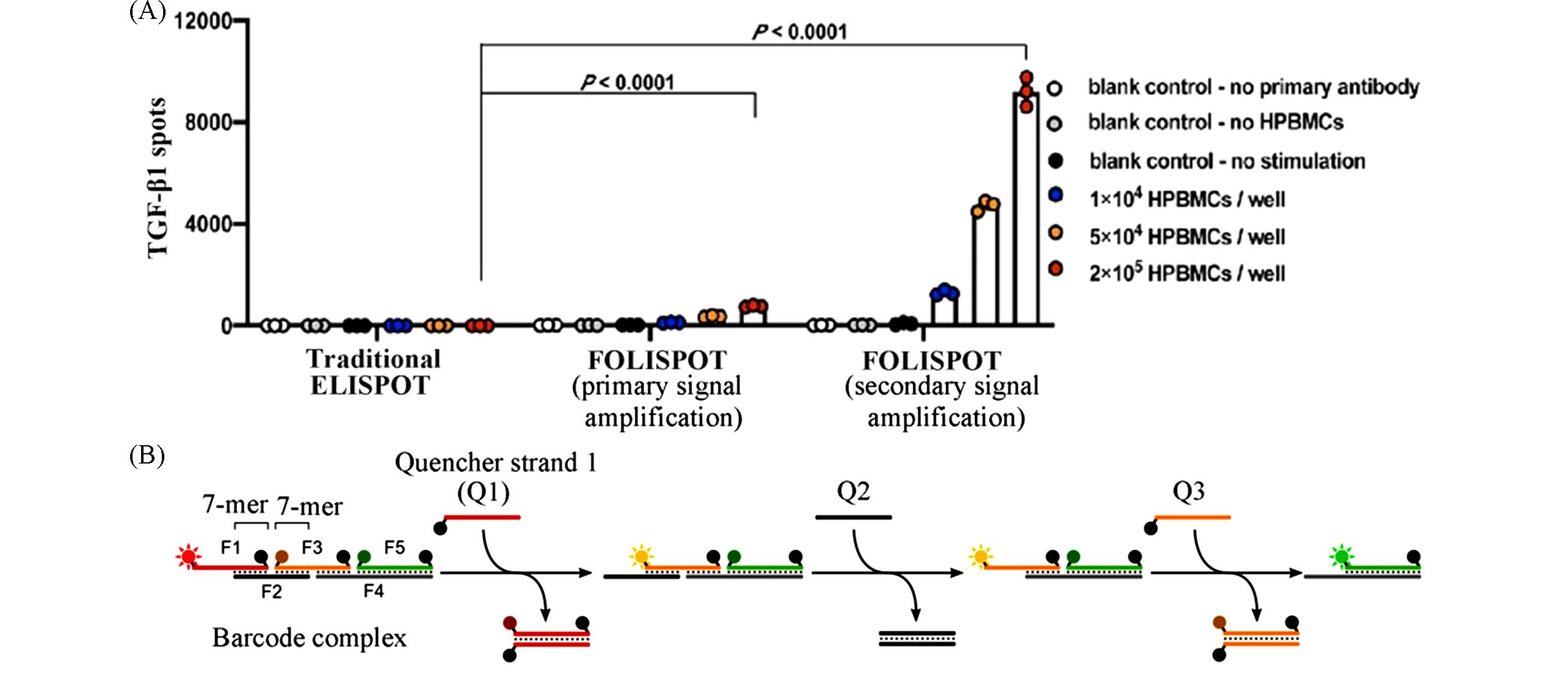
Fig.9 Data and schematic of FOLISPOT and CCFB(A) Data of the comparison between ELISpot, primary and secondary signal amplification FOLISPOT[60]; (B) schematic of CCFB based on strand displacement reaction[61].(A) Copyright 2022, American Chemical Society; (B) Copyright 2022, American Chemical Society.
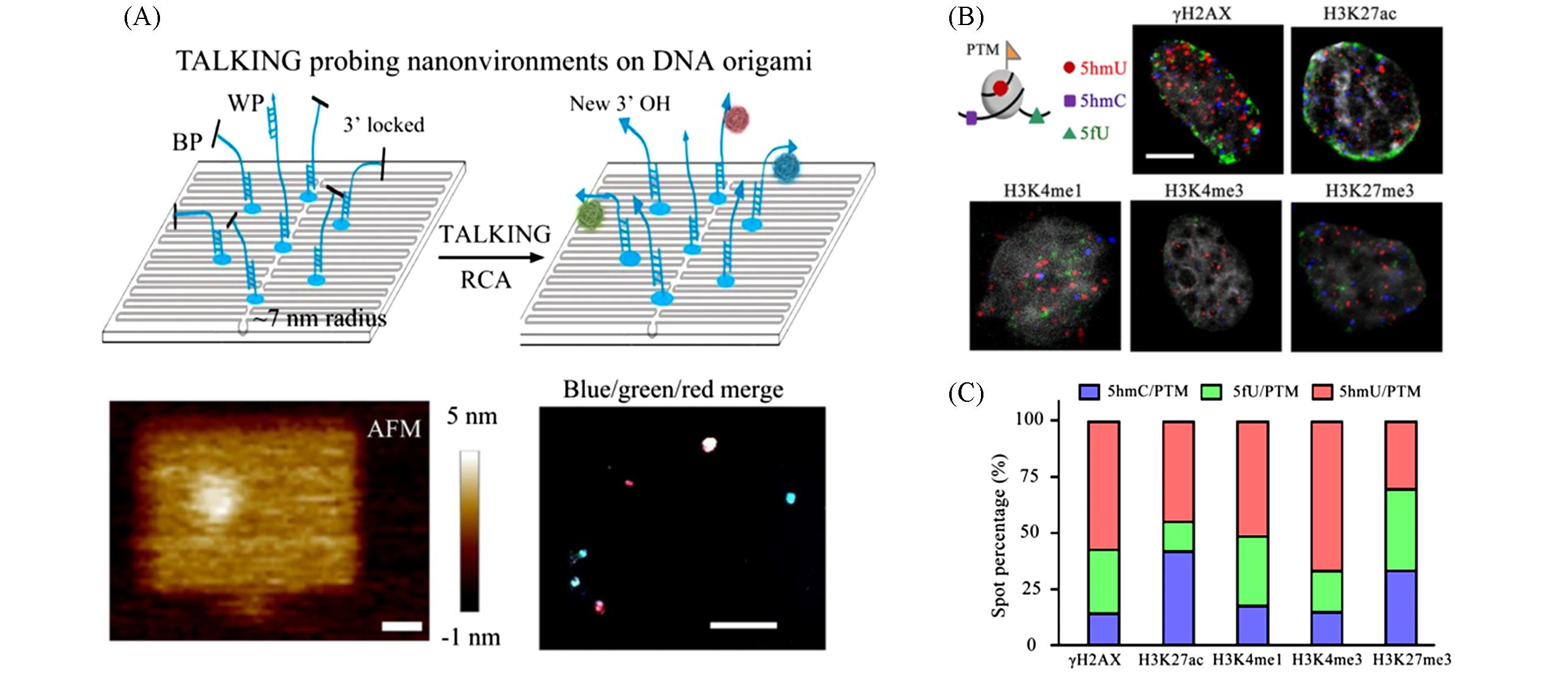
Fig.10 Schematic and data of Cell⁃TALKING[22](A) Schematic of Cell-TALKING on DNA origami substrates; WP and BP represent histone and surrounding chromatin modifications, respectively; (B) merged cell images for five histone PTMs; (C) the spot percentages of three encoded chromatin modifications of single cells in five samples.Copyright 2021, Springer Nature.
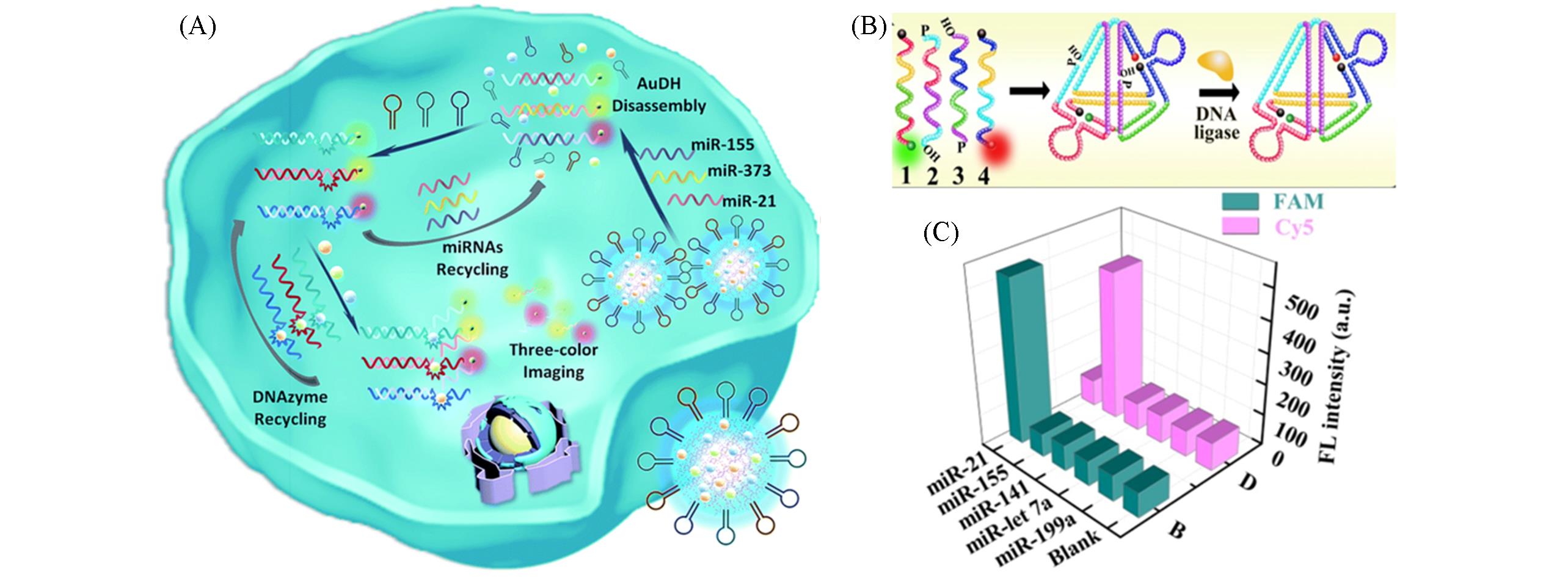
Fig.11 Schematic and data of nanoparticles and DNA nanostructures encoded amplification methods in live cells(A) Schematic of AuDH drived miRNA imaging[78]; (B) structure of multicolor-encoded DNA nanostructures[80]; (C) specificity evaluation of the DNA TetrNano probes for the target miRNA-21 and miRNA-155[80].(A) Copyright 2018, the authors. (B, C) Copyright 2016, American Chemical Society.
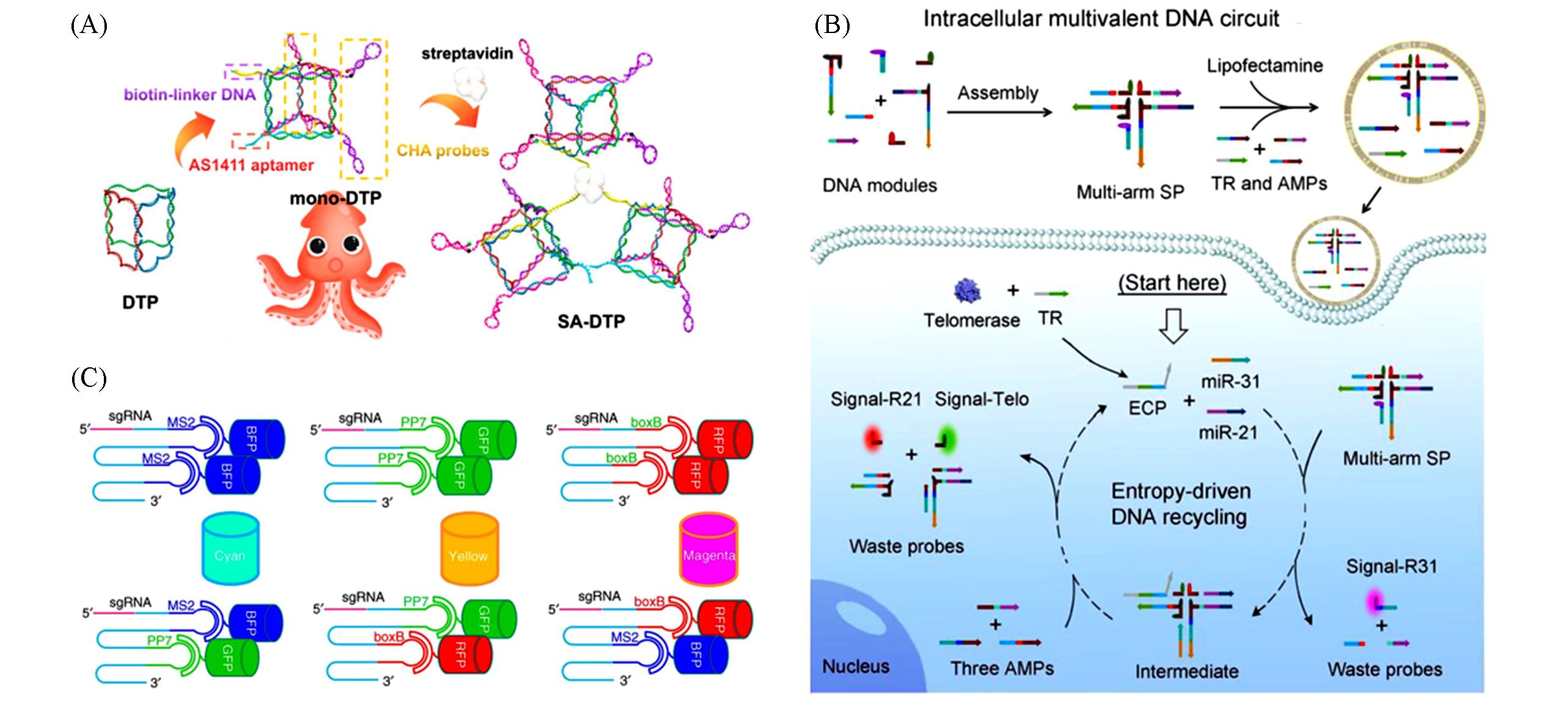
Fig.12 Structure of DNA nanostructures and circuits and schematic of CRISPRainbow(A) Structure of SA-DTP[81]; (B) schematic of intracellular entropy-driven multivalent DNA circuits[69]; (C) schematic of CRISPRainbow for multiplexed labeling of genomic loci[83].(A) Copyright 2020, American Chemical Society; (B) Copyright 2020, Wiley-VCH; (C) Copyright 2016, Springer Nature.
| Method | Principle | Nucleic acid amplification | Target | Encoding number | Ref. |
|---|---|---|---|---|---|
| MERFISH | Hybridization reaction | None | mRNA | 100—1000 | [ |
| Improved MERFISH | Hybridization reaction | bDNA | mRNA | 130 | [ |
| SeqFISH | Hybridization reaction | None | mRNA | 12 per cycle | [ |
| HCR⁃seqFISH | Hybridization reaction | smHCR | mRNA | 250 | [ |
| STARmap | Hybridization reaction | RCA | mRNA | 160—1020 | [ |
| SABER⁃FISH | Hybridization reaction | PER | mRNA | 17 per cycle | [ |
| SeqEA | Hybridization reaction | RCA | mRNA | 36 per cycle | [ |
| Clicker⁃FISH | Coupling reaction | RCA | RNA structures and polyadenylation | 3 per cycle | [ |
| Sc5hmU/5hmC⁃microgel | Coupling reaction | RCA | 5hmU and 5hmC | 2 per cycle | [ |
| CODEX | Antigen⁃antibody reaction | None | Proteins | 2 per cycle | [ |
| IsHCR | Antigen⁃antibody reaction | HCR | Proteins | 3 per cycle | [ |
| ELISpot | Antigen⁃antibody reaction | PER | Proteins | 6 per cycle | [ |
| CCFB | Antigen⁃antibody reaction | None | Proteins | 3 per cycle | [ |
| Cell⁃TALKING | Combinational reaction | RCA | 5hmC, 5hmU and 5fU | 3 per cycle | [ |
| ClampFISH | Combinational reaction | Click⁃amplifying | mRNA | 3 per cycle | [ |
Table 1 Properties of in situ nucleic acids-encoded amplification methods
| Method | Principle | Nucleic acid amplification | Target | Encoding number | Ref. |
|---|---|---|---|---|---|
| MERFISH | Hybridization reaction | None | mRNA | 100—1000 | [ |
| Improved MERFISH | Hybridization reaction | bDNA | mRNA | 130 | [ |
| SeqFISH | Hybridization reaction | None | mRNA | 12 per cycle | [ |
| HCR⁃seqFISH | Hybridization reaction | smHCR | mRNA | 250 | [ |
| STARmap | Hybridization reaction | RCA | mRNA | 160—1020 | [ |
| SABER⁃FISH | Hybridization reaction | PER | mRNA | 17 per cycle | [ |
| SeqEA | Hybridization reaction | RCA | mRNA | 36 per cycle | [ |
| Clicker⁃FISH | Coupling reaction | RCA | RNA structures and polyadenylation | 3 per cycle | [ |
| Sc5hmU/5hmC⁃microgel | Coupling reaction | RCA | 5hmU and 5hmC | 2 per cycle | [ |
| CODEX | Antigen⁃antibody reaction | None | Proteins | 2 per cycle | [ |
| IsHCR | Antigen⁃antibody reaction | HCR | Proteins | 3 per cycle | [ |
| ELISpot | Antigen⁃antibody reaction | PER | Proteins | 6 per cycle | [ |
| CCFB | Antigen⁃antibody reaction | None | Proteins | 3 per cycle | [ |
| Cell⁃TALKING | Combinational reaction | RCA | 5hmC, 5hmU and 5fU | 3 per cycle | [ |
| ClampFISH | Combinational reaction | Click⁃amplifying | mRNA | 3 per cycle | [ |
| 1 | Allam M., Cai S., Coskun A. F., NPJ Precis. Oncol., 2020, 4, 11 |
| 2 | Jia Y. M., Lou X. D., Xia F., Chinese J. Anal. Chem., 2018, 46(9), 1329—1338 |
| 贾永梅, 娄筱叮, 夏帆. 分析化学, 2018, 46(9), 1329—1338 | |
| 3 | Schwartzman O., Tanay A., Nat. Rev. Genet., 2015, 16(12), 716—726 |
| 4 | Noureen N., Wu S., Lv Y., Yang J., Alfred Yung W. K., Gelfond J., Wang X., Koul D., Ludlow A., Zheng S., Nat. Commun., 2021, 12(1), 139 |
| 5 | Sungkaworn T., Jobin M. L., Burnecki K., Weron A., Lohse M. J., Calebiro D., Nature, 2017, 550(7677), 543—547 |
| 6 | Ma Y., Wang M., Li W., Zhang Z., Zhang X., Tan T., Zhang X. E., Cui Z., Nat. Commun., 2017, 8(1), 15318 |
| 7 | Saito K., Chang Y. F., Horikawa K., Hatsugai N., Higuchi Y., Hashida M., Yoshida Y., Matsuda T., Arai Y., Nagai T., Nat. Commun., 2012, 3(1), 1262 |
| 8 | Liu Y., Lu Y., Yang X., Zheng X., Wen S., Wang F., Vidal X., Zhao J., Liu D., Zhou Z., Ma C., Zhou J., Piper J. A., Xi P., Jin D., Nature, 2017, 543(7644), 229—233 |
| 9 | Grimm J. B., English B. P., Chen J., Slaughter J. P., Zhang Z., Revyakin A., Patel R., Macklin J. J., Normanno D., Singer R. H., Lionnet T., Lavis L. D., Nat. Methods, 2015, 12(3), 244—250 |
| 10 | Ebrahimi S. B., Samanta D., Mirkin C. A., J. Am. Chem. Soc., 2020, 142(26), 11343—11356 |
| 11 | Luo F., Qin G., Xia T., Fang X., Annu. Rev. Anal. Chem.(Palo. Alto. Calif.), 2020, 13(1), 337—361 |
| 12 | Su X., Xiao X., Zhang C., Zhao M., Appl. Spectrosc., 2012, 66(11), 1249—1261 |
| 13 | Cao X., Yu H., Xue J., Bai M., Zhao Y., Li Y., Zhao Y., Chen F., Anal. Chem., 2020, 92(13), 9356—9361 |
| 14 | Qin P., Parlak M., Kuscu C., Bandaria J., Mir M., Szlachta K., Singh R., Darzacq X., Yildiz A., Adli M., Nat. Commun., 2017, 8(1), 14725 |
| 15 | Brenner S., Lerner R. A., Proceedings of the National Academy of Sciences, 1992, 89(12), 5381—5383 |
| 16 | Clark M. A., Acharya R. A., Arico⁃Muendel C. C., Belyanskaya S. L., Benjamin D. R., Carlson N. R., Centrella P. A., Chiu C. H., Creaser S. P., Cuozzo J. W., Davie C. P., Ding Y., Franklin G. J., Franzen K. D., Gefter M. L., Hale S. P., Hansen N. J. V., Israel D. I., Jiang J., Kavarana M. J., Kelley M. S., Kollmann C. S., Li F., Lind K., Mataruse S., Medeiros P. F., Messer J. A., Myers P., O'keefe H., Oliff M. C., Rise C. E., Satz A. L., Skinner S. R., Svendsen J. L., Tang L., van Vloten K., Wagner R. W., Yao G., Zhao B., Morgan B. A., Nat. Chem. Biol., 2009, 5(9), 647—654 |
| 17 | Cai B., Kim D., Akhand S., Sun Y., Cassell R. J., Alpsoy A., Dykhuizen E. C., van Rijn R. M., Wendt M. K., Krusemark C. J., J. Am. Chem. Soc., 2019, 141(43), 17057—17061 |
| 18 | Ståhl P. L., Salmén F., Vickovic S., Lundmark A., Navarro J. F., Magnusson J., Giacomello S., Asp M., Westholm J. O., Huss M., Mollbrink A., Linnarsson S., Codeluppi S., Borg Å., Pontén F., Costea P. I., Sahlén P., Mulder J., Bergmann O., Lundeberg J., Frisén J., Science, 2016, 353(6294), 78—82 |
| 19 | Boettiger A. N., Bintu B., Moffitt J. R., Wang S., Beliveau B. J., Fudenberg G., Imakaev M., Mirny L. A., Wu C. T., Zhuang X., Nature, 2016, 529(7586), 418—422 |
| 20 | Zhuang X., Nat. Methods, 2021, 18(1), 18—22 |
| 21 | Kress W. J., Erickson D. L., Proceed. Nat. Acad. Sciences, 2008, 105(8), 2761—2762 |
| 22 | Chen F., Bai M., Cao X., Xue J., Zhao Y., Wu N., Wang L., Zhang D., Zhao Y., Nat. Commun., 2021, 12(1), 1965 |
| 23 | Chen F., Xue J., Zhang J., Bai M., Yu X., Fan C., Zhao Y., J. Am. Chem. Soc., 2020, 142(6), 2889—2896 |
| 24 | Abramson R. D., Myers T. W., Curr. Opin. Biotechnol., 1993, 4(1), 41—47 |
| 25 | Zhao Y., Chen F., Li Q., Wang L., Fan C., Chem. Rev., 2015, 115(22), 12491—12545 |
| 26 | Kishi J. Y., Schaus T. E., Gopalkrishnan N., Xuan F., Yin P., Nat. Chem., 2018, 10(2), 155—164 |
| 27 | Zhou W., Li D., Yuan R., Xiang Y., Anal. Chem., 2019, 91(5), 3628—3635 |
| 28 | Friedrich M. W., Curr. Opin. Biotechnol., 2006, 17(1), 59—66 |
| 29 | Sahagun⁃Ruiz A., Waghela S. D., Holman P. J., Chieves L. P., Wagner G. G., Veter. Parasit., 1997, 73(1), 53—63 |
| 30 | Chen S. X., Zhang D. Y., Seelig G., Nat. Chem., 2013, 5(9), 782—789 |
| 31 | Kishi J. Y., Beliveau B. J., Lapan S. W., West E. R., Zhu A., Sasaki H. M., Saka S. K., Wang Y., Cepko C. L., Yin P., bioRxiv, 2018, 401810 |
| 32 | Takei Y., Yun J., Zheng S., Ollikainen N., Pierson N., White J., Shah S., Thomassie J., Suo S., Eng C. H. L., Guttman M., Yuan G. C., Cai L., Nature, 2021, 590(7845), 344—350 |
| 33 | Crosetto N., Bienko M., van Oudenaarden A., Nat. Rev. Genet., 2015, 16(1), 57—66 |
| 34 | Femino A. M., Fay F. S., Fogarty K., Singer R. H., Science, 1998, 280(5363), 585—590 |
| 35 | Fan Y., Braut S. A., Lin Q., Singer R. H., Skoultchi A. I., Genomics, 2001, 71(1), 66—69 |
| 36 | Lubeck E., Cai L., Nat. Methods, 2012, 9(7), 743—748 |
| 37 | Chen K. H., Boettiger A. N., Moffitt J. R., Wang S., Zhuang X., Science, 2015, 348(6233), aaa6090 |
| 38 | Moffitt J. R., Hao J., Bambah⁃Mukku D., Lu T., Dulac C., Zhuang X., Proc. Nat. Acad. Sci. USA, 2016, 113(50), 14456—14461 |
| 39 | Emanuel G., Moffitt J. R., Zhuang X., Nat. Methods, 2017, 14(12), 1159—1162 |
| 40 | Zhang M., Eichhorn S. W., Zingg B., Yao Z., Cotter K., Zeng H., Dong H., Zhuang X., Nature, 2021, 598(7879), 137—143 |
| 41 | Xia C., Babcock H. P., Moffitt J. R., Zhuang X., Scientific Reports, 2019, 9(1), 7721 |
| 42 | Lubeck E., Coskun A. F., Zhiyentayev T., Ahmad M., Cai L., Nat. Methods, 2014, 11(4), 360—361 |
| 43 | Shah S., Lubeck E., Zhou W., Cai L., Neuron, 2016, 92(2), 342—357 |
| 44 | Wang X., Allen W. E., Wright M. A., Sylwestrak E. L., Samusik N., Vesuna S., Evans K., Liu C., Ramakrishnan C., Liu J., Nolan G. P., Bava F. A., Deisseroth K., Science, 2018, 361(6400), eaat5691 |
| 45 | Kishi J. Y., Lapan S. W., Beliveau B. J., West E. R., Zhu A., Sasaki H. M., Saka S. K., Wang Y., Cepko C. L., Yin P., Nat. Methods, 2019, 16(6), 533—544 |
| 46 | Deng R., Zhang K., Wang L., Ren X., Sun Y., Li J., Chem., 2018, 4(6), 1373—1386 |
| 47 | Kleiner R. E., Dumelin C. E., Liu D. R., Chem. Soc. Rev., 2011, 40(12), 5707—5717 |
| 48 | Neri D., Lerner R. A., Annu. Rev. Biochem., 2018, 87(1), 479—502 |
| 49 | Rouhanifard S. H., Mellis I. A., Dunagin M., Bayatpour S., Jiang C. L., Dardani I., Symmons O., Emert B., Torre E., Cote A., Sullivan A., Stamatoyannopoulos J. A., Raj A., Nat. Biotechnol., 2019, 37(1), 84—89 |
| 50 | Besanceney⁃Webler C., Jiang H., Zheng T., Feng L., Soriano Del Amo D., Wang W., Klivansky L. M., Marlow F. L., Liu Y., Wu P., Angew. Chem. Int. Ed., 2011, 50(35), 8051—8056 |
| 51 | Cañeque T., Müller S., Rodriguez R., Nat. Rev. Chem., 2018, 2(9), 202—215 |
| 52 | Raulf A., Spahn C. K., Zessin P. J. M., Finan K., Bernhardt S., Heckel A., Heilemann M., RSC Adv., 2014, 4(57), 30462—30466 |
| 53 | Chen F., Bai M., Cao X., Zhao Y., Xue J., Zhao Y., Nucleic Acids Res., 2019, 47(22), e145 |
| 54 | Bai M., Cao X., Chen F., Xue J., Zhao Y., Zhao Y., Anal. Chem., 2021, 93(30), 10495—10501 |
| 55 | Agasti S. S., Liong M., Peterson V. M., Lee H., Weissleder R., J. Am. Chem. Soc., 2012, 134(45), 18499—18502 |
| 56 | Agasti S. S., Wang Y., Schueder F., Sukumar A., Jungmann R., Yin P., Chem. Sci., 2017, 8(4), 3080—3091 |
| 57 | Wang Y., Woehrstein J. B., Donoghue N., Dai M., Avendaño M. S., Schackmann R. C. J., Zoeller J. J., Wang S. S. H., Tillberg P. W., Park D., Lapan S. W., Boyden E. S., Brugge J. S., Kaeser P. S., Church G. M., Agasti S. S., Jungmann R., Yin P., Nano Letters, 2017, 17(10), 6131—6139 |
| 58 | Goltsev Y., Samusik N., Kennedy⁃Darling J., Bhate S., Hale M., Vazquez G., Black S., Nolan G. P., Cell, 2018, 174(4), 968— 981(e15) |
| 59 | Lin R., Feng Q., Li P., Zhou P., Wang R., Liu Z., Wang Z., Qi X., Tang N., Shao F., Luo M., Nat. Methods, 2018, 15(4), 275—278 |
| 60 | Ma J., Peng Z., Ma L., Diao L., Shao X., Zhao Z., Liu L., Zhang L., Huang C., Liu M., Anal. Chem., 2022, 94(24), 8704—8714 |
| 61 | Makino K., Susaki E. A., Endo M., Asanuma H., Kashida H., J. Am. Chem. Soc., 2022, 144(4), 1572—1579 |
| 62 | Murayama K., Kamiya Y., Kashida H., Asanuma H., ChemBioChem, 2015, 16(9), 1298—1301 |
| 63 | Xue J., Chen F., Su L., Cao X., Bai M., Zhao Y., Fan C., Zhao Y., Angew. Chem. Int. Ed., 2021, 60(7), 3428—3432 |
| 64 | Zhang K., Deng R., Teng X., Li Y., Sun Y., Ren X., Li J., J. Am. Chem. Soc., 2018, 140(36), 11293—11301 |
| 65 | Hattori N., Niwa T., Kimura K., Helin K., Ushijima T., Nucleic Acids Res., 2013, 41(15), 7231—7239 |
| 66 | Xue J., Chen F., Bai M., Cao X., Huang P., Zhao Y., Anal. Chem., 2019, 91(7), 4696—4701 |
| 67 | Zhang J., Zhao P., Li W., Ye L., Li L., Li Z., Li M., Angew. Chem. Int. Ed., 2022, 61(22), e202117562 |
| 68 | Wang Z., Niu J., Zhao C., Wang X., Ren J., Qu X., Angew. Chem. Int. Ed., 2021, 60(22), 12431—12437 |
| 69 | Bai M., Chen F., Cao X., Zhao Y., Xue J., Yu X., Fan C., Zhao Y., Angew. Chem. Int. Ed., 2020, 59(32), 13267—13272 |
| 70 | Kang J. H., Jang W. Y., Ko Y. T., Pharm. Res., 2017, 34(4), 704—717 |
| 71 | Sun L., Gao Y., Wang Y., Wei Q., Shi J., Chen N., Li D., Fan C., Chem. Sci., 2018, 9(27), 5967—5975 |
| 72 | St. Martin A., Salamango D., Serebrenik A., Shaban N., Brown W. L., Donati F., Munagala U., Conticello S. G., Harris R. S., Nucleic Acids Res., 2018, 46(14), e84 |
| 73 | Stewart J. A., Schauer G., Bhagwat A. S., Nucleic Acids Res., 2020, 48(20), e118 |
| 74 | Schweissthal B., Brunken K., Brach J., Emde L., Hetsch F., Fricke S., Meier J. C., bioRxiv, 2021, 2021.03.03.433736 |
| 75 | Wang J., Yu S., Wu Q., Gong X., He S., Shang J., Liu X., Wang F., Angew. Chem. Int. Ed., 2021, 60(19), 10766—10774 |
| 76 | Wei J., Wang H., Wu Q., Gong X., Ma K., Liu X., Wang F., Angew. Chem. Int. Ed., 2020, 59(15), 5965—5971 |
| 77 | Liu C., Chen Y., Zhao J., Wang Y., Shao Y., Gu Z., Li L., Zhao Y., Angew. Chem. Int. Ed., 2021, 60(26), 14324—14328 |
| 78 | Meng X., Zhang K., Dai W., Cao Y., Yang F., Dong H., Zhang X., Chem. Sci., 2018, 9(37), 7419—7425 |
| 79 | Yuan P., Mao X., Liew S. S., Wu S., Huang Y., Li L., Yao S. Q., ACS Appl. Mater. Interfaces, 2020, 12(52), 57695—57709 |
| 80 | Zhou W., Li D., Xiong C., Yuan R., Xiang Y., ACS Appl. Mater. Interfaces, 2016, 8(21), 13303—13308 |
| 81 | Yang F., Cheng Y., Zhang Y., Wei W., Dong H., Lu H., Zhang X., Anal. Chem., 2020, 92(6), 4411—4418 |
| 82 | Lu H., Guo K., Cao Y., Yang F., Wang D., Dou L., Liu Y., Dong H., Anal. Chem., 2020, 92(2), 1850—1855 |
| 83 | Ma H., Tu L. C., Naseri A., Huisman M., Zhang S., Grunwald D., Pederson T., Nat. Biotechnol., 2016, 34(5), 528—530 |
| 84 | Ma H., Tu L. C., Naseri A., Chung Y. C., Grunwald D., Zhang S., Pederson T., Nat. Methods, 2018, 15(11), 928—931 |
| 85 | Askary A., Sanchez⁃Guardado L., Linton J. M., Chadly D. M., Budde M. W., Cai L., Lois C., Elowitz M. B., Nat. Biotechnol., 2020, 38(1), 66—75 |
| 86 | Frieda K. L., Linton J. M., Hormoz S., Choi J., Chow K. H. K., Singer Z. S., Budde M. W., Elowitz M. B., Cai L., Nature, 2017, 541(7635), 107—111 |
| 87 | Takei Y., Shah S., Harvey S., Qi L. S., Cai L., Biophys. J., 2017, 112(9), 1773—1776 |
| [1] | 汪诗琪, 罗博文, 俞计成, 顾臻. 近红外二区活体荧光成像在肿瘤诊疗中的应用[J]. 高等学校化学学报, 2022, 43(12): 20220577. |
| [2] | 马小飞, 胡山, 李俊彬, 杨盛, 谌委菊, 卿志和, 周怡波, 杨荣华. 细胞内源性分子辅助荧光信号放大策略及细胞成像[J]. 高等学校化学学报, 2022, 43(12): 20220320. |
| [3] | 陈尚钰, 沈清明, 孙鹏飞, 范曲立. 小分子基温敏聚合物纳米粒子的制备及在近红外二区荧光成像与光热治疗中的应用[J]. 高等学校化学学报, 2022, 43(12): 20220392. |
| [4] | 常通航, 程震. 整合荧光成像和化疗的有机小分子诊疗探针的研究进展[J]. 高等学校化学学报, 2022, 43(12): 20220430. |
| [5] | 张钤, 刘雅薇, 王帆, 刘凯, 张洪杰. 稀土纳米材料在高分辨活体成像及诊疗中的应用[J]. 高等学校化学学报, 2022, 43(12): 20220552. |
| [6] | 刘苗, 刘瑞波, 刘巴蒂, 钱鹰. 溶酶体靶向吲哚氟硼二吡咯光敏剂的合成、 双光子荧光成像及光动力治疗[J]. 高等学校化学学报, 2022, 43(10): 20220326. |
| [7] | 陈宏达, 张婳, 王振新. 用于小动物活体的荧光-光热双模成像系统[J]. 高等学校化学学报, 2021, 42(3): 725. |
| [8] | 王萌萌, 栾天骄, 杨铭焱, 吕佳佳, 高杰, 李洪玉, 卫钢, 袁泽利. 肿瘤乏氧靶向响应的罗丹明荧光探针及其成像介导手术治疗[J]. 高等学校化学学报, 2021, 42(10): 3071. |
| [9] | 梁钰昕, 赵容, 梁馨月, 方晓红. 细胞膜上信号转导蛋白的单分子成像与分析[J]. 高等学校化学学报, 2020, 41(6): 1127. |
| [10] | 白翠婷, 岳仁叶, 罗列高, 马楠. 基于双色荧光传感器的癌细胞成像及microRNA定量检测[J]. 高等学校化学学报, 2020, 41(6): 1252. |
| [11] | 邵伟, LEE Jiyoung, 李方园, 凌代舜. 有机小分子纳米粒子的光学诊疗应用[J]. 高等学校化学学报, 2020, 41(11): 2356. |
| [12] | 张勇,申城,幸志荣,陈归柒,卢资,侯志兵,陈雪梅. 可视化检测次氯酸的苯并咪唑类荧光增强型探针[J]. 高等学校化学学报, 2019, 40(12): 2480. |
| [13] | 刘晔, 姚顺雨, 方超, 赵外欧, 王静媛, 李亚鹏. 靶向髓过氧化酶的双模态探针分子的合成与表征[J]. 高等学校化学学报, 2018, 39(7): 1573. |
| [14] | 张涛, 汤永嘉, 徐亮, 刘克良. 新型可用于荧光标记的α-氰基丙烯酸酯单体的合成及在小鼠活体成像中的应用[J]. 高等学校化学学报, 2016, 37(6): 1168. |
| [15] | 米小龙, 焦晓洁, 刘畅, 何松, 曾宪顺. 基于罗丹明荧光信号报告基团的细胞通透性铜离子荧光探针[J]. 高等学校化学学报, 2016, 37(10): 1784. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||