

高等学校化学学报 ›› 2019, Vol. 40 ›› Issue (2): 288.doi: 10.7503/cjcu20180653
许炎, 刘翠( ), 韩成娟, 潘明玉, 孙照琦, 韩冰玉, 杨忠志
), 韩成娟, 潘明玉, 孙照琦, 韩冰玉, 杨忠志
收稿日期:2018-09-26
出版日期:2019-02-10
发布日期:2018-11-19
作者简介:联系人简介: 刘 翠, 女, 博士, 讲师, 主要从事理论与计算化学研究. E-mail:
基金资助:
XU Yan, LIU Cui*( ), HAN Chengjuan, PAN Mingyu, SUN Zhaoqi, HAN Bingyu, YANG Zhongzhi
), HAN Chengjuan, PAN Mingyu, SUN Zhaoqi, HAN Bingyu, YANG Zhongzhi
Received:2018-09-26
Online:2019-02-10
Published:2018-11-19
Contact:
LIU Cui
E-mail:liuc@lnnu.edu.cn
Supported by:摘要:
发展了应用于鸟嘌呤G和氨基酸残基体系的浮动电荷力场, 该力场明确定义了孤对电子和键的电荷和位置, 通过电荷随着环境的浮动来体现极化效应; 通过氢键拟合函数kHB描绘了氢键键能. 应用量子化学方法, 对G与氨基酸残基体系从氢键、 几何结构及电荷分布3个方面展开计算及分析, 并以其为基准, 确定参数发展了适用于G与氨基酸残基氢键体系的ABEEMσπ PFF. 采用3种不同力场模拟目标分子的结构和性质. 模拟结果表明, 发展的ABEEMσπ PFF与量子化学方法具有最好的一致性, 可用于模拟生物大分子体系.
中图分类号:
TrendMD:
许炎, 刘翠, 韩成娟, 潘明玉, 孙照琦, 韩冰玉, 杨忠志. 鸟嘌呤与氨基酸残基体系的极化力场. 高等学校化学学报, 2019, 40(2): 288.
XU Yan,LIU Cui,HAN Chengjuan,PAN Mingyu,SUN Zhaoqi,HAN Bingyu,YANG Zhongzhi. Development of Polarization Force Field for Guanine and Amino Acid Residues Systems†. Chem. J. Chinese Universities, 2019, 40(2): 288.
| Hydrogen bond | k' | C | rlp,H | D |
|---|---|---|---|---|
| lpN307102, H-N101106 | 0.7010 | 0.0931 | 1.3019 | 0.0319 |
| lpN307102, H-N101109 | 0.5438 | 0.0931 | 1.3019 | 0.0319 |
| lpN307102, H-N101111 | 0.5840 | 0.0931 | 1.3019 | 0.0319 |
| lpN307202, H-N101111 | 0.5744 | 0.2500 | 2.0000 | 0.0500 |
| lpO308102, H-N101116 | 0.5800 | 0.0931 | 1.5019 | 0.0319 |
| lpO308102, H-N101117 | 0.5450 | 0.0931 | 1.3019 | 0.0319 |
| lpO308201, H-N101106 | 0.6000 | 0.0792 | 2.0874 | 0.3202 |
| lpO308201, H-N101116 | 0.5850 | 0.0506 | 1.2523 | 0.0536 |
| lpO308201, H-N101117 | 0.6030 | 0.0931 | 1.3019 | 0.0319 |
| lpO308207, H-N101106 | 0.4800 | 0.0931 | 1.3019 | 0.0319 |
| lpO308207, H-N101111 | 0.6570 | 0.0951 | 1.5019 | 0.0519 |
| lpS316102, H-N101116 | 0.8000 | 0.0931 | 1.3019 | 0.0319 |
| lpS316102, H-N101117 | 0.6400 | 0.0931 | 1.3019 | 0.0319 |
Table 1 Parameters k', C, rlp,H and D in the function kHB(Rlp,H)
| Hydrogen bond | k' | C | rlp,H | D |
|---|---|---|---|---|
| lpN307102, H-N101106 | 0.7010 | 0.0931 | 1.3019 | 0.0319 |
| lpN307102, H-N101109 | 0.5438 | 0.0931 | 1.3019 | 0.0319 |
| lpN307102, H-N101111 | 0.5840 | 0.0931 | 1.3019 | 0.0319 |
| lpN307202, H-N101111 | 0.5744 | 0.2500 | 2.0000 | 0.0500 |
| lpO308102, H-N101116 | 0.5800 | 0.0931 | 1.5019 | 0.0319 |
| lpO308102, H-N101117 | 0.5450 | 0.0931 | 1.3019 | 0.0319 |
| lpO308201, H-N101106 | 0.6000 | 0.0792 | 2.0874 | 0.3202 |
| lpO308201, H-N101116 | 0.5850 | 0.0506 | 1.2523 | 0.0536 |
| lpO308201, H-N101117 | 0.6030 | 0.0931 | 1.3019 | 0.0319 |
| lpO308207, H-N101106 | 0.4800 | 0.0931 | 1.3019 | 0.0319 |
| lpO308207, H-N101111 | 0.6570 | 0.0951 | 1.5019 | 0.0519 |
| lpS316102, H-N101116 | 0.8000 | 0.0931 | 1.3019 | 0.0319 |
| lpS316102, H-N101117 | 0.6400 | 0.0931 | 1.3019 | 0.0319 |
| Class | V2/(kJ·mol-1) | Class | V2/(kJ·mol-1) | Class | V2/(kJ·mol-1) |
|---|---|---|---|---|---|
| 84-71-75-75 | 6.90 | 83-71-79-81 | 5.65 | 75-73-76-71 | 41.84 |
| 84-71-75-72 | 6.90 | 78-72-75-75 | 17.36 | 75-73-76-82 | 41.84 |
| 84-71-76-73 | 7.11 | 75-72-78-71 | 20.08 | 83-74-78-71 | 10.04 |
| 84-71-76-82 | 7.11 | 75-72-78-74 | 20.08 | 83-74-78-72 | 10.04 |
| 78-71-79-75 | 5.65 | 76-73-75-75 | 10.67 | 71-75-75-73 | 22.80 |
| 83-71-79-71 | 5.65 | 76-73-75-79 | 6.28 | 72-75-75-73 | 22.80 |
Table 2 Dihedral angle parameters
| Class | V2/(kJ·mol-1) | Class | V2/(kJ·mol-1) | Class | V2/(kJ·mol-1) |
|---|---|---|---|---|---|
| 84-71-75-75 | 6.90 | 83-71-79-81 | 5.65 | 75-73-76-71 | 41.84 |
| 84-71-75-72 | 6.90 | 78-72-75-75 | 17.36 | 75-73-76-82 | 41.84 |
| 84-71-76-73 | 7.11 | 75-72-78-71 | 20.08 | 83-74-78-71 | 10.04 |
| 84-71-76-82 | 7.11 | 75-72-78-74 | 20.08 | 83-74-78-72 | 10.04 |
| 78-71-79-75 | 5.65 | 76-73-75-75 | 10.67 | 71-75-75-73 | 22.80 |
| 83-71-79-71 | 5.65 | 76-73-75-79 | 6.28 | 72-75-75-73 | 22.80 |
| Class | ν/(kJ·mol-1) | Class | ν/(kJ·mol-1) | Class | ν/(kJ·mol-1) | Class | ν/(kJ·mol-1) |
|---|---|---|---|---|---|---|---|
| 72-75-75-71 | 41.84 | 71-72-79-81 | 41.84 | 79-78-71-83 | 41.84 | 75-75-71-84 | 41.84 |
| 79-75-75-71 | 41.84 | 71-75-79-81 | 41.84 | 79-79-71-83 | 41.84 | 76-75-71-84 | 41.84 |
| 79-75-75-73 | 41.84 | 73-71-76-82 | 41.84 | 78-83-74-83 | 87.86 | 79-75-71-84 | 41.84 |
| 71-71-79-81 | 41.84 | 79-75-71-83 | 41.84 |
Table 3 Improper dihedral angle parameters
| Class | ν/(kJ·mol-1) | Class | ν/(kJ·mol-1) | Class | ν/(kJ·mol-1) | Class | ν/(kJ·mol-1) |
|---|---|---|---|---|---|---|---|
| 72-75-75-71 | 41.84 | 71-72-79-81 | 41.84 | 79-78-71-83 | 41.84 | 75-75-71-84 | 41.84 |
| 79-75-75-71 | 41.84 | 71-75-79-81 | 41.84 | 79-79-71-83 | 41.84 | 76-75-71-84 | 41.84 |
| 79-75-75-73 | 41.84 | 73-71-76-82 | 41.84 | 78-83-74-83 | 87.86 | 79-75-71-84 | 41.84 |
| 71-71-79-81 | 41.84 | 79-75-71-83 | 41.84 |
| X—H…Y | Sum | LX—Y/nm | LH—Y/nm | θ/(°) | X—H…Y | Sum | LX—Y/nm | LH—Y/nm | θ/(°) |
|---|---|---|---|---|---|---|---|---|---|
| O…HN | 13 | 0.28 | 0.19 | 158.9 | S…HN | 2 | 0.34 | 0.25 | 154.8 |
| N…HN | 13 | 0.29 | 0.21 | 150.2 | O…HS | 1 | 0.34 | 0.24 | 123.2 |
| O…HO | 1 | 0.27 | 0.19 | 142.8 | N…HS | 1 | 0.34 | 0.23 | 131.7 |
| N…HO | 1 | 0.28 | 0.20 | 144.4 |
Table 4 Average hydrogen bond length(L) and hydrogen bond angle(θ) of complexes
| X—H…Y | Sum | LX—Y/nm | LH—Y/nm | θ/(°) | X—H…Y | Sum | LX—Y/nm | LH—Y/nm | θ/(°) |
|---|---|---|---|---|---|---|---|---|---|
| O…HN | 13 | 0.28 | 0.19 | 158.9 | S…HN | 2 | 0.34 | 0.25 | 154.8 |
| N…HN | 13 | 0.29 | 0.21 | 150.2 | O…HS | 1 | 0.34 | 0.24 | 123.2 |
| O…HO | 1 | 0.27 | 0.19 | 142.8 | N…HS | 1 | 0.34 | 0.23 | 131.7 |
| N…HO | 1 | 0.28 | 0.20 | 144.4 |
| Complex | Charge/e | ||||||
|---|---|---|---|---|---|---|---|
| N6 | O9 | N10 | H11 | N13 | H15 | N16 | |
| G3Arg1 | -0.046* | 0.012 | 0.012 | 0.018 | -0.045* | 0.022 | 0.013 |
| G3Arg2 | -0.048* | 0.012 | 0.012 | 0.020 | -0.046* | 0.021 | 0.014 |
| G4Arg1 | 0.017 | 0.008 | 0.009 | -0.057* | 0.041 | 0.019 | -0.015 |
| G4Arg2 | 0.017 | 0.008 | 0.009 | -0.057* | 0.042 | 0.019 | -0.015 |
| G5Arg1 | -0.037 | 0.014 | 0.012 | 0.019 | -0.072* | 0.024 | 0.014 |
| G5Arg2 | -0.038 | 0.013 | 0.012 | 0.019 | -0.071* | 0.024 | 0.014 |
| G6Arg1 | -0.070* | 0.008 | 0.011 | 0.016 | -0.050 | 0.016 | 0.014 |
| G6Arg2 | -0.070* | 0.008 | 0.012 | 0.016 | -0.046 | 0.016 | 0.013 |
| G3Lys | -0.060* | 0.017 | 0.013 | 0.023 | -0.055* | 0.028 | 0.021 |
| G1Asn1 | 0.002 | 0.001 | -0.011* | -0.021* | 0.002 | 0.001 | 0.034* |
| G2Asn1 | 0 | -0.018* | 0.003 | -0.003 | -0.036* | 0.041* | -0.010 |
| G3Asn2 | -0.015* | 0.003 | 0.012 | 0.008 | -0.001* | 0.007 | -0.008 |
| G1Ser | 0.002 | 0.002 | -0.003* | -0.017* | 0.005 | 0.004 | 0.019* |
| G2Ser | 0.002 | -0.011* | 0.011 | 0.006 | -0.025* | 0.033* | -0.010 |
| G1Cys | 0.001 | 0.001 | 0.006* | -0.002* | 0.004 | 0.003 | -0.004 |
| G2Cys | 0.001 | -0.004* | 0.011 | 0.005 | -0.012* | 0.009* | -0.010 |
Table 5 Change of atomic charges in isolated base G and hydrogen bonded complexes
| Complex | Charge/e | ||||||
|---|---|---|---|---|---|---|---|
| N6 | O9 | N10 | H11 | N13 | H15 | N16 | |
| G3Arg1 | -0.046* | 0.012 | 0.012 | 0.018 | -0.045* | 0.022 | 0.013 |
| G3Arg2 | -0.048* | 0.012 | 0.012 | 0.020 | -0.046* | 0.021 | 0.014 |
| G4Arg1 | 0.017 | 0.008 | 0.009 | -0.057* | 0.041 | 0.019 | -0.015 |
| G4Arg2 | 0.017 | 0.008 | 0.009 | -0.057* | 0.042 | 0.019 | -0.015 |
| G5Arg1 | -0.037 | 0.014 | 0.012 | 0.019 | -0.072* | 0.024 | 0.014 |
| G5Arg2 | -0.038 | 0.013 | 0.012 | 0.019 | -0.071* | 0.024 | 0.014 |
| G6Arg1 | -0.070* | 0.008 | 0.011 | 0.016 | -0.050 | 0.016 | 0.014 |
| G6Arg2 | -0.070* | 0.008 | 0.012 | 0.016 | -0.046 | 0.016 | 0.013 |
| G3Lys | -0.060* | 0.017 | 0.013 | 0.023 | -0.055* | 0.028 | 0.021 |
| G1Asn1 | 0.002 | 0.001 | -0.011* | -0.021* | 0.002 | 0.001 | 0.034* |
| G2Asn1 | 0 | -0.018* | 0.003 | -0.003 | -0.036* | 0.041* | -0.010 |
| G3Asn2 | -0.015* | 0.003 | 0.012 | 0.008 | -0.001* | 0.007 | -0.008 |
| G1Ser | 0.002 | 0.002 | -0.003* | -0.017* | 0.005 | 0.004 | 0.019* |
| G2Ser | 0.002 | -0.011* | 0.011 | 0.006 | -0.025* | 0.033* | -0.010 |
| G1Cys | 0.001 | 0.001 | 0.006* | -0.002* | 0.004 | 0.003 | -0.004 |
| G2Cys | 0.001 | -0.004* | 0.011 | 0.005 | -0.012* | 0.009* | -0.010 |
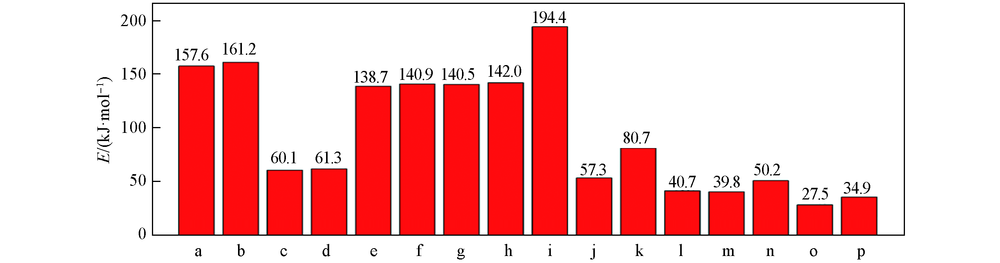
Fig.5 Hydrogen bond energies of hydrogen bond complexesa. G3Arg1; b. G3Arg2; c. G4Arg1; d. G4Arg2; e. G5Arg1; f. G5Arg2; g. G6Arg1; h. G6Arg2; i. G3Lys; j. G1Asn1; k. G2Asn1; l. G3Asn2; m. G1Ser; n. G2Ser; o. G1Cys; p. G2Cys.
| Species | ABEEMσπ PFF | OPLS/AA | AMBER OL15 |
|---|---|---|---|
| Bond length/nm | 0.0015 | 0.0019 | 0.0019 |
| Angle/(°) | 1.6 | 2.2 | 1.9 |
| Torsion/(°) | 2.3 | 3.0 | 2.8 |
Table 6 RMSDs of geometric structure between force fields and M06-2X/6-311++G** method
| Species | ABEEMσπ PFF | OPLS/AA | AMBER OL15 |
|---|---|---|---|
| Bond length/nm | 0.0015 | 0.0019 | 0.0019 |
| Angle/(°) | 1.6 | 2.2 | 1.9 |
| Torsion/(°) | 2.3 | 3.0 | 2.8 |
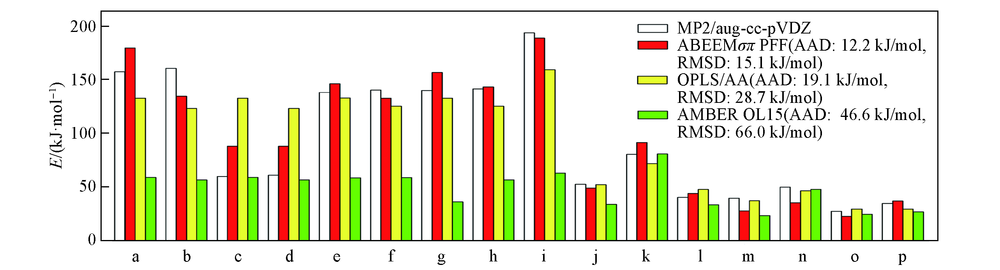
Fig.8 Hydrogen bond energies of QM and various force fieldsa. G3Arg1; b. G3Arg2; c. G4Arg1; d. G4Arg2; e. G5Arg1; f. G5Arg2; g. G6Arg1; h. G6Arg2; i. G3Lys; j. G1Asn1; k. G2Asn1; l. G3Asn2; m. G1Ser; n. G2Ser; o. G1Cys; p. G2Cys.
| Complex | Time/s | Complex | Time/s | ||||||
|---|---|---|---|---|---|---|---|---|---|
| MP2/ aug-cc-pVDZ | ABEEMσπ PFF | OPLS/AA | AMBER OL15 | MP2/ aug-cc-pVDZ | ABEEMσπ PFF | OPLS/AA | AMBER OL15 | ||
| G3Arg1 | 30739.2 | 1.1 | 1.0 | 1.0 | G1Lys | 42499.2 | 1.2 | 1.0 | 1.0 |
| G3Arg2 | 28354.2 | 1.0 | 0.9 | 0.9 | G1Asn1 | 22165.8 | 1.0 | 0.9 | 0.9 |
| G4Arg1 | 33514.8 | 1.1 | 1.0 | 1.0 | G2Asn1 | 21605.4 | 1.0 | 0.9 | 0.9 |
| G4Arg2 | 33964.8 | 1.1 | 1.0 | 1.0 | G3Asn2 | 34574.4 | 1.0 | 0.9 | 0.9 |
| G5Arg1 | 32125.8 | 1.0 | 0.9 | 0.9 | G1Ser | 15585.6 | 1.0 | 0.9 | 0.9 |
| G5Arg2 | 33345.2 | 1.1 | 1.0 | 1.0 | G2Ser | 17013.6 | 1.0 | 0.9 | 0.9 |
| G6Arg1 | 33349.7 | 1.1 | 0.9 | 0.9 | G1Cys | 15991.2 | 1.0 | 0.9 | 0.9 |
| G6Arg2 | 33764.9 | 1.0 | 0.9 | 0.9 | G2Cys | 15226.2 | 1.0 | 0.9 | 0.9 |
Table 7 Computational time of single point energy by various methods
| Complex | Time/s | Complex | Time/s | ||||||
|---|---|---|---|---|---|---|---|---|---|
| MP2/ aug-cc-pVDZ | ABEEMσπ PFF | OPLS/AA | AMBER OL15 | MP2/ aug-cc-pVDZ | ABEEMσπ PFF | OPLS/AA | AMBER OL15 | ||
| G3Arg1 | 30739.2 | 1.1 | 1.0 | 1.0 | G1Lys | 42499.2 | 1.2 | 1.0 | 1.0 |
| G3Arg2 | 28354.2 | 1.0 | 0.9 | 0.9 | G1Asn1 | 22165.8 | 1.0 | 0.9 | 0.9 |
| G4Arg1 | 33514.8 | 1.1 | 1.0 | 1.0 | G2Asn1 | 21605.4 | 1.0 | 0.9 | 0.9 |
| G4Arg2 | 33964.8 | 1.1 | 1.0 | 1.0 | G3Asn2 | 34574.4 | 1.0 | 0.9 | 0.9 |
| G5Arg1 | 32125.8 | 1.0 | 0.9 | 0.9 | G1Ser | 15585.6 | 1.0 | 0.9 | 0.9 |
| G5Arg2 | 33345.2 | 1.1 | 1.0 | 1.0 | G2Ser | 17013.6 | 1.0 | 0.9 | 0.9 |
| G6Arg1 | 33349.7 | 1.1 | 0.9 | 0.9 | G1Cys | 15991.2 | 1.0 | 0.9 | 0.9 |
| G6Arg2 | 33764.9 | 1.0 | 0.9 | 0.9 | G2Cys | 15226.2 | 1.0 | 0.9 | 0.9 |
| [1] | Douki T., Berger M., Raoul S., Ravanat J.L., Cadet J., Determination of 8-Oxo-Purines in DNA by HPLC Using Amperometric Detection., Birkhäuser Press, Basel, 1995, 213-224 |
| [2] | Manas E. S., Getahun Z., Wright W. W., DeGradoW. F., Vanderkooi J. M., J. Am. Chem. Soc.,2000, 122(41), 9883-9890 |
| [3] | Shukla A., Barbiellini B., Buslaps T., Suortti P. Z., Phys. Chem.,2001, 215(10), 1315-1321 |
| [4] | Cadet J., Douki T., Ravanat J. L., Photochem. Photobio.,2015, 91(1), 140-155 |
| [5] | Huo H. J., Zhao D. X., Yang Z. Z., Chem. J. Chinese Universities,2011, 32(12), 2877-2884 |
| (霍红杰, 赵东霞, 杨忠志. 高等学校化学学报, 2011, 32(12), 2877-2884) | |
| [6] | Liu C., Zhang Q. H., Gong L. D., Lu L. N., Yang Z. Z., Chem. J. Chinese Universities,2014, 35(12), 2645-2653 |
| (刘翠, 张千慧, 宫利东, 卢丽男, 杨忠志. 高等学校化学学报, 2014, 35(12), 2645-2653 | |
| [7] | Liu C., Li Y., Han B. Y., Gong L. D., Lu L. N., Yang Z. Z., Zhao D. X., J. Chem. Theory Comput.,2017, 13(5), 2098-2111 |
| [8] | Zhang Q. H., Wang Y., Liu C., Yang Z. Z., Acta Chim. Sinica,2014, 72(8), 956-962 |
| (张千慧, 王阳, 刘翠, 杨忠志. 化学学报, 2014, 72(8), 956-962) | |
| [9] | Liu S., Li S. S., Liu D. J., Wang C. S., Acta Phys. Chim. Sin., 2013, 29(12), 2551-2557 |
| (刘硕, 李书实, 刘冬佳, 王长生. 物理化学学报, 2013, 29(12), 2551-2557) | |
| [10] | Wang C. S., Liu P., Yu N., Acta Phys. Chim. Sin., 2013, 29(6), 1173-1182 |
| (王长生, 刘朋, 于楠. 物理化学学报, 2013, 29(6), 1173-1182) | |
| [11] | González J., Baños I., León I., Contreras-Garcia J., Cocinero E. J., Lesarri A., Fernández J. A., Millán J., J. Chem. Theory Comput., 2016, 12(2), 523-534 |
| [12] | Xie W., Gao J., J. Chem. Theory Comput.,2007, 3(6), 1890-1900 |
| [13] | Wang J., Cieplak P., Li J., Cai Q., Hsieh M., Luo R., Duan Y., J. Phys. Chem. B,2012, 116(24), 7088-7101 |
| [14] | Ivani I., Dans P. D., Noy A., Perez A., Faustino L., Hospital A., Walther J., Andrio P., Goni R., Balaceanu A., Portella G., Battistini F., Gelpi J. L., Gonzalez C., Vendruscolo M., Laughton C. A., Harris S. A., Case D. A., Orozco M., Nat. Methods,2016, 13(1), 55-58 |
| [15] | Kaminski G. A., Stern H. A., Berne B. J., Friesner R. A., Cao Y. X., Murphy R. B., Zhou R., Halgren T. A., J. Comput. Chem., 2002, 23(16), 1515-1531 |
| [16] | Jorgensen W. L., Jensen K. P., Alexandrova A. N., J. Chem. Theory Comput., 2007, 3(6), 1987-1992 |
| [17] | Savelyev A., MacKerell J. A. D., J. Phys. Chem. B,2014, 118(24), 6742-6757 |
| [18] | Lemkul J. A., Huang J., Roux B., Mackerell A. D. Jr., Chem. Rev., 2016, 116(9), 4983-5013 |
| [19] | Zhang C., Bell D., Harger M., Ren P. Y., J. Chem. Theory Comput., 2017, 13(2), 666-678 |
| [20] | Donchev A. G., Ozrin V. D., Subbotin M. V., Tarasov O. V., Tarasov V. I. A., Proc. Natl. Acad. Sci. U. S. A., 2005, 102(22), 7829-7834 |
| [21] | Donchev A. G., Galkin N. G., Illarionov A. A., Khoruzhii O. V., Olevanov M. A., Ozrin V. D., Subbotin M. V., Tarasov V. I., Proc. Natl. Acad. Sci. U. S. A. ,2006, 103(23), 8613-8617 |
| [22] | Zhao D. X., Liu C., Wang F. F., Yu C. Y., Gong L. D., Liu S. B., Yang Z. Z., J. Chem. Theory Comput., 2010, 6(3), 795-804 |
| [23] | Wu Y., Yang Z. Z., J. Phys. Chem. A,2004, 108(37), 7563-7576 |
| [24] | Li X., Yang Z. Z., J. Phys. Chem. A,2005, 109(18), 4102-4111 |
| [25] | Yang Z. Z., Wang J. J., Zhao D. X., J. Comput. Chem., 2014, 35(23), 1690-1706 |
| [26] | Frisch M.J., Trucks G. W., Schlegel H. B., Scuseria G. E., Robb M. A., Cheeseman J. R., Scalmani G., Barone V., Mennucci B., Petersson G. A., Nakatsuji H., Caricato M., Li X., Hratchian H. P., Izmaylov A. F., Bloino J., Zheng G., Sonnenberg J. L., Hada M., Ehara M., Toyota K., Fukuda R., Hasegawa J., Ishida M., Nakajima T., Honda Y., Kitao O., Nakai H., Vreven T., Montgomery J. A., Peralta J. E., Ogliaro F., Bearpark M., Heyd J. J., Brothers E., Kudin K. N., Staroverov V. N., Kobayashi R., Normand J., Raghavachari K., Rendell A., Burant J. C., Iyengar S. S., Tomasi J., Cossi M., Rega N., Millam J. M., Klene M., Knox J. E., Cross J. B., Bakken V., Adamo C., Jaramillo J., Gomperts R., Stratmann R. E., Yazyev O., Austin A. J., Cammi R., Pomelli C., Ochterski J. W., Martin R. L., Morokuma K., Zakrzewski V. G., Voth G. A., Salvador P., Dannenberg J. J., Dapprich S., Daniels A. D., Farkas O., Foresman J. B., Ortiz J. V., Cioslowski J., Fox D. J., Gaussian 09, Revision A.02, Gaussian Inc., Wallingford CT, 2009 |
| [27] | Hohenstein E. G., Chill S. T., Sherrill C. D., J. Chem. Theory Comput., 2008, 4(12), 1996-2000 |
| [1] | 陈佳璐 黄硕. 纳米孔测序技术在核酸修饰检测中的应用[J]. 高等学校化学学报, 2022, 43(Album-4): 20220333. |
| [2] | 季双琦, 靳钊, 观文娜, 潘翔宇, 关彤. 双阳离子型离子液体和十八烷基修饰的混合模式硅胶固定相的制备及色谱性能[J]. 高等学校化学学报, 2022, 43(6): 20220008. |
| [3] | 闵婧, 王力彦. 利用三中心氢键限制芳酰胺构象的核磁共振氢谱分析[J]. 高等学校化学学报, 2022, 43(6): 20220084. |
| [4] | 张勇, 许俊, 鲍雨, 崔树勋. 非极性有机溶剂对分子内氢键弱化程度的单分子力谱定量研究[J]. 高等学校化学学报, 2022, 43(4): 20210863. |
| [5] | 崔韶丽, 张维佳, 邵学广, 蔡文生. 自由能计算揭示苏氨酸对抗冻蛋白与冰晶结合能力的影响[J]. 高等学校化学学报, 2022, 43(3): 20210838. |
| [6] | 胡波, 朱昊辰. 双层氧化石墨烯纳米体系中受限水的介电常数[J]. 高等学校化学学报, 2022, 43(2): 20210614. |
| [7] | 高慧玲, 曹珍珍, 顾芳, 王海军. 氢键型水凝胶自修复行为的Monte Carlo模拟[J]. 高等学校化学学报, 2022, 43(11): 20220482. |
| [8] | 彭沁, 方业广, 张腾烁, 崔刚龙, 方维海. DNA环境中硒代胸腺嘧啶和腺嘌呤碱基对的激发态性质和光物理机理的理论研究[J]. 高等学校化学学报, 2021, 42(7): 2136. |
| [9] | 王乐, 秦刘磊, 刘洋, 任丽, 徐慧婷, 刘尊奇. 一维链状氢键型甘氨酸超分子化合物[(Gly)2+ (18-crown-6)2(MnCl4)2‒]的合成、 结构及介电性质[J]. 高等学校化学学报, 2021, 42(3): 691. |
| [10] | 倪卿盛, 杜淼, 单国荣, 宋义虎, 吴子良, 郑强. 一维粒子对聚乙烯醇水溶液流变行为的调控[J]. 高等学校化学学报, 2021, 42(12): 3738. |
| [11] | 李晓蕾, 孙云娇, 唐颖, 王长生. 醇及脱氧核糖与水分子间三体作用强度的快速准确计算[J]. 高等学校化学学报, 2021, 42(12): 3664. |
| [12] | 巩珊珊, 吴彤, 王官格, 黄擎, 苏岳锋, 吴锋. 基于高效回收废旧锂离子电池正极材料的低共熔溶剂的筛选[J]. 高等学校化学学报, 2021, 42(10): 3151. |
| [13] | 白兰, 翟磊, 王畅鸥, 何民辉, 莫松, 范琳. 含酰胺结构超低膨胀聚酰亚胺薄膜的热膨胀行为[J]. 高等学校化学学报, 2020, 41(4): 795. |
| [14] | 秦刘磊,刘洋,关小琴,郑晓媛,张子钰,刘尊奇. 无机-有机杂化化合物[(H2DABCO)CuCl4]·H2O的合成及开关型介电性质[J]. 高等学校化学学报, 2020, 41(1): 70. |
| [15] | 徐宇, 花儿. 烷基乙二胺-CF3CO2型质子化离子液体的分子间氢键作用[J]. 高等学校化学学报, 2018, 39(9): 1954. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||