

高等学校化学学报 ›› 2020, Vol. 41 ›› Issue (5): 872.doi: 10.7503/cjcu20190684
• 庆祝《高等学校化学学报》复刊40周年专栏 • 上一篇 下一篇
李文帅1,3,武国瑞2,3,张茜菁3,岳爱琴2,杜维俊2,赵晋忠1,*( ),刘定斌3,*(
),刘定斌3,*( )
)
收稿日期:2019-12-17
出版日期:2020-05-10
发布日期:2020-02-26
通讯作者:
赵晋忠,刘定斌
E-mail:zhaojinzhongnd@126.com;liudb@nankai.edu.cn
基金资助:
LI Wenshuai1,3,WU Guorui2,3,ZHANG Xijing3,YUE Aiqin2,DU Weijun2,ZHAO Jinzhong1,*( ),LIU Dingbin3,*(
),LIU Dingbin3,*( )
)
Received:2019-12-17
Online:2020-05-10
Published:2020-02-26
Contact:
Jinzhong ZHAO,Dingbin LIU
E-mail:zhaojinzhongnd@126.com;liudb@nankai.edu.cn
Supported by:摘要:
细菌是一种与人类生命活动息息相关的微生物, 其快速、 高灵敏检测对重大传染性疾病的防控至关重要. 本文介绍了拉曼光谱用于细菌检测的基本原理, 综述了3种拉曼光谱用于细菌检测的主要方式, 包括细菌组成成分检测、 细菌代谢物检测以及基于拉曼探针标记的检测模式, 并对各种拉曼检测方法进行了分析比较. 最后, 展望了拉曼光谱在细菌检测领域的发展前景, 并提出了5条建议.
中图分类号:
TrendMD:
李文帅, 武国瑞, 张茜菁, 岳爱琴, 杜维俊, 赵晋忠, 刘定斌. 基于拉曼光谱的细菌检测研究进展. 高等学校化学学报, 2020, 41(5): 872.
LI Wenshuai, WU Guorui, ZHANG Xijing, YUE Aiqin, DU Weijun, ZHAO Jinzhong, LIU Dingbin. Advances in Bacterial Detection Based on Raman Spectroscopy †. Chem. J. Chinese Universities, 2020, 41(5): 872.
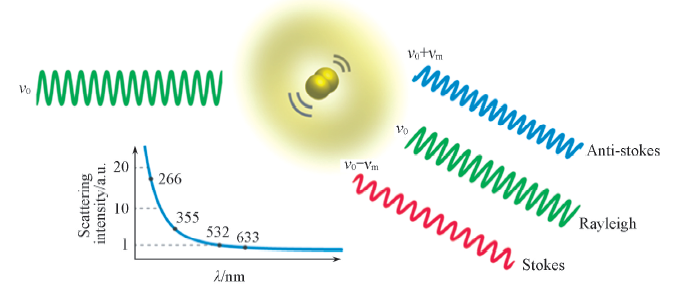
Fig.1 Rayleigh(elastic) and Raman(inelastic) scattering at a molecule, and scattering intensity dependence on the wavelength of the scattered light(for ease of comparison, the scattering intensity is arbitrarily set to 1 at a wavelength of 532 nm)[7] Copyright 2013, Annual Reviews.
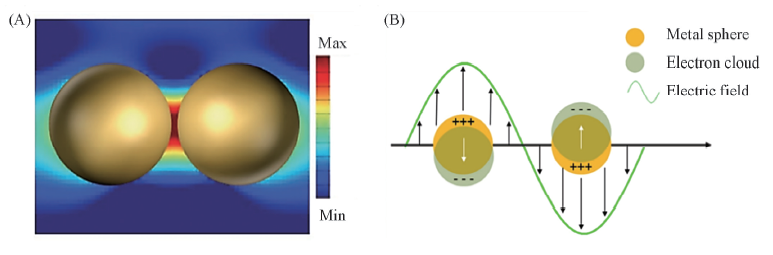
Fig.2 Representative electric field distribution of SERS-active Au nanoparticles Note that the SERS “hot-spot” is generated in the gap between two closely arranged nanoparticles(A)[9], and collective oscillations of electrons in a spherical nanoparticle under the action of the external electric field(B)[10]. (A) Copyright 2018, Wiley-VCH;(B) Copyright 2017, MDPI.
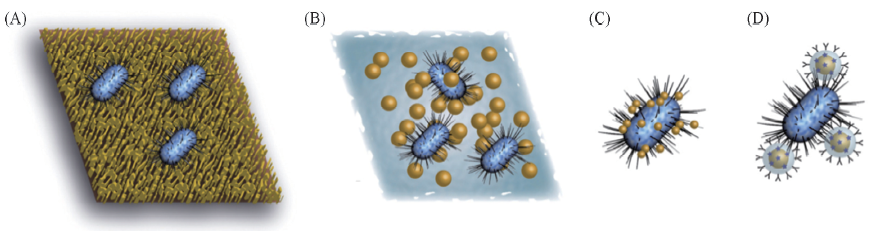
Fig.5 Schematic display of different approaches for SERS based detection of bacteria Whole cells can be measured on a solid substrate(A) or in a colloid suspension(B) and it is also possible to attach or deposit metallic nanoparticles directly on the bacterial cell wall(C) or to apply with SERS tags(D)[52] Copyright 2015, Elsevier.
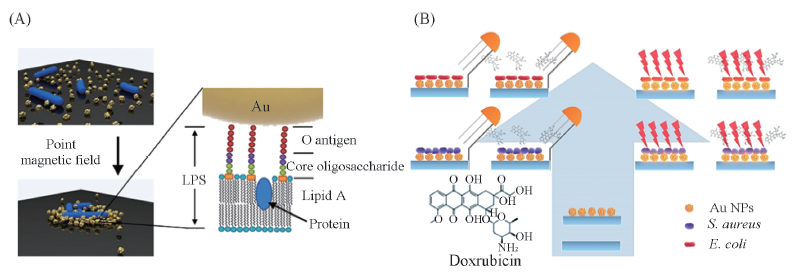
Fig.6 Schematics of the condensation process of Au-MNPs and bacteria(left) and the biomolecular characteristics of the bacterial cell wall that can possibly be detected by SERS(right)(A)[54] and schematic illustration for bacteria detection by using nanostructured Au based biosensor(B)[55] (A) Copyright 2019, Elsevier; (B) Copyright 2014, American Chemical Society.
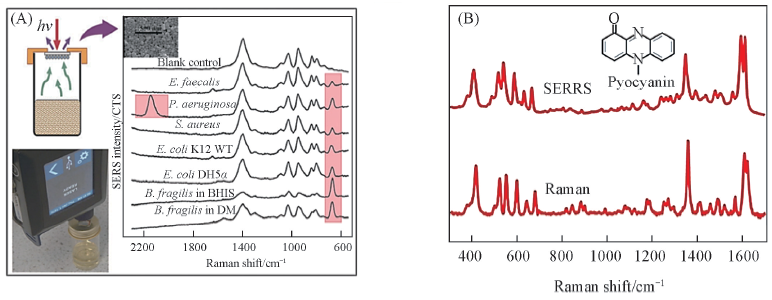
Fig.7 SERS spectra of the headspace(A)[64] and Raman and SERS spectra of pyocyainn(B)[65] (A) Copyright 2018, John Wiley and Sons Ltd.; (B) Copyright 2016, Springer Nature.
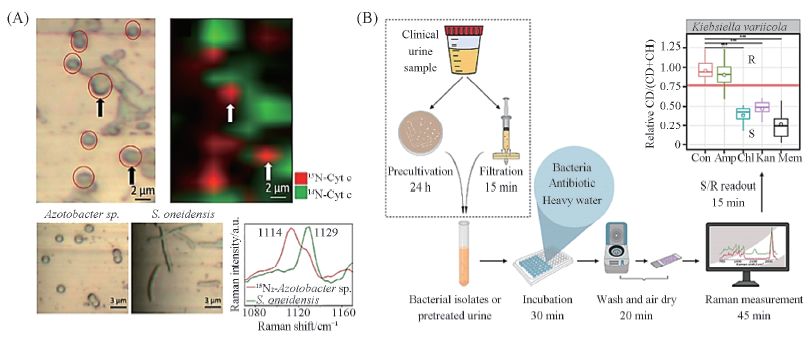
Fig.8 Probing and Raman imaging of N2-fixing bacteria in artfcial communities(A)[67] and flow chart of rapid Raman drug sensitivity test based on heavy water labeling(B)[68] (A) Copyright 2018, American Chemical Society; (B) Copyright 2019, American Chemical Society.
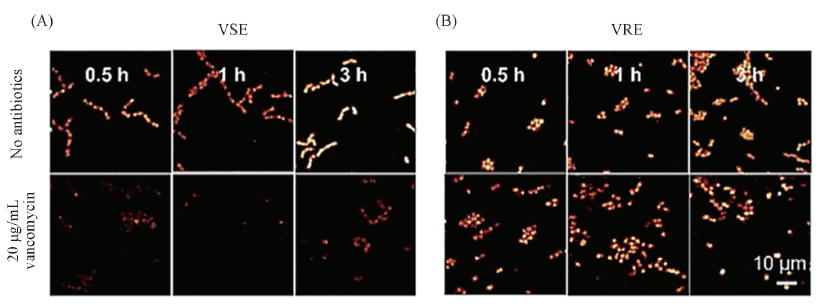
Fig.9 Time lapse of C-D component concentration map in bacteria cultivated in glucose-d7 medium C-D component concentration map in VSE(A) and VRE(B), in the presence and absence of 20 μg/mL vancomycin[70]. Copyright 2018, American Chemical Society.
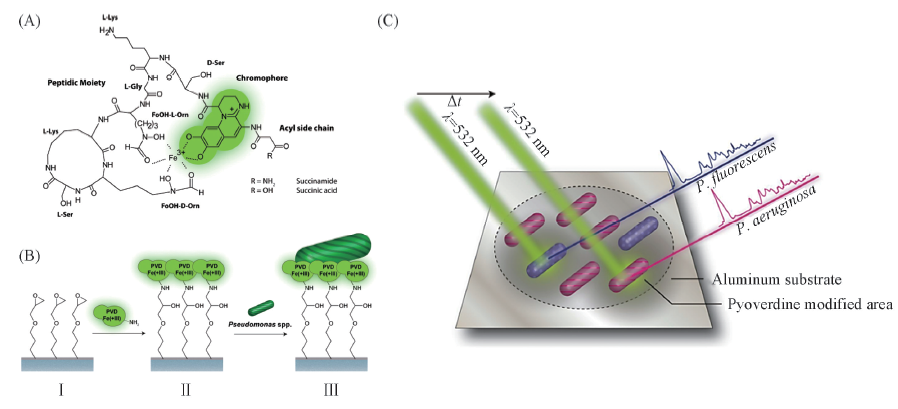
Fig.11 Structure of the pyoverdine iron complex of P. fluorescens DSM 50090(A), scheme illustrating the proposed isolation strategy for Pseudomonas spp.(B) and Raman detection of P. fluorescens and P. aeruginosa(C)[84] (B) The (3-glycidyloxypropyl)-trimethoxysilane modified surface(Ⅰ) is functionalized with an pyoverdine iron complex(Ⅱ), subsequently the chip can be used for capturing various Pseudomonas species(Ⅲ). Copyright 2016, American Chemical Society.

Fig.12 Schematic illustration of the synthesis of Au-Van SERS tags(A), the synthesis of aptamer-modified Fe3O4@AuMNPs(B), and the operating procedure for S. aureus detection via the dual-recognition SERS biosensor(C)[85] Copyright 2019, Elsevier.

Fig.13 Phage specifically recognizes the bacteria and then immobilizes the bacteria to the surface of a nano substrate with SERS activity, able to get extremely strong Raman signals[87] Copyright 2017, American Chemical Society.
| [1] |
Kuramochi H., Takeuchi S., Yonezawa K., Kamikubo H., Kataoka M., Tahara T., Nat. Chem, 2017,9(7), 660—666
doi: 10.1038/nchem.2717 URL pmid: 28644485 |
| [2] |
Bergholt M. S., Albro M. B., Stevens M. M., Biomaterials., 2017,140(3), 128—137
doi: 10.1016/j.biomaterials.2017.06.015 URL |
| [3] | Grousd J. A., Rich H. E., Alcorn J. F., Clin. Microbiol. Rev., 2019,32(3), 99—103 |
| [4] | Tsalik E. L., Bonomo R. A., Fowler V. G., Annu. Rev. Med., 2018,69(4), 379—394 |
| [5] |
Piano S., Bartoletti M., Tonon M., Baldassarre M., Chies G., Romano A., Viale P., Vettore E., Domenicali M., Stanco M., Pilutti C., Frigo A. C., Brocca A., Bernardi M., Caraceni P., Angeli P., Gut, 2018,67(10), 1892—1899
doi: 10.1136/gutjnl-2017-314324 URL pmid: 28860348 |
| [6] |
Pang X., Xiao Q., Cheng Y., Ren E., Lian L., Zhang Y., Gao H., Wang X., Leung W., Chen X., Liu G., Xu C ., ACS Nano., 2019,13(2), 2427—2438
doi: 10.1021/acsnano.8b09336 URL pmid: 30657302 |
| [7] | Opilik L., Schmid T., Zenobi R., Annu. Rev. Anal. Chem., 2013,6(1), 379—398 |
| [8] | Fleischmann M., Hendra P. J., Mcquillan A. J., Chem. Phys. Lett., 1974,26(2), 163—166 |
| [9] | Galvan D. D., Yu Q., Adv. Healthc. Mater., 2018,7(13), 36—63 |
| [10] | Wang J. C., Lin W. H., Cao E., Xu X. F., Liang W. J., Zhang X. F., Sensors-Basel., 2017,17(12), 2719—2738 |
| [11] |
Kumar N., Weckhuysen B. M., Wain A. J., Pollard A. J., Nat. Protoc., 2019,14(4), 1169—1193
doi: 10.1038/s41596-019-0132-z URL pmid: 30911174 |
| [12] |
Yilmaz M., Babur E., Ozdemir M., Gieseking R. L., Dede Y., Tamer U., Schatz G. C., Facchetti A., Usta H., Demirel G., Nat. Mater., 2017,16(9), 918—924
doi: 10.1038/nmat4957 URL pmid: 28783157 |
| [13] | Kühn M., Ivleva N. P., Klitzke S., Niessner R., Baumann T., Sci. Total. Environ., 2015,535(5), 122—130 |
| [14] | Georgiou R., Gueriau P., Sahle C. J., Bernard S., Mirone A., Garrouste R., Bergmann U., Rueff J. P., Bertrand L., Sci. Adv., 2019,5(8), 496—502 |
| [15] |
Jiang Z., Gao P., Yang L., Huang C., Li Y., Anal. Chem, 2015,87(24), 12177—12182
doi: 10.1021/acs.analchem.5b03058 URL pmid: 26575213 |
| [16] |
Chen Z., Jiang S., Kang G., Nguyen D., Schatz G. C., Duyne R. P., J. Am. Chem. Soc., 2019,141(39), 15684—15692
doi: 10.1021/jacs.9b07979 URL pmid: 31503482 |
| [17] | Pakapongpan S., Poo-Arporn R. P., Mat. Sci. End. C-Mater., 2017,76(5), 398—405 |
| [18] |
Keidel A., López I., Staffa J., Kuhlmann U., Bozoglian F., Gimbert-Suriñach C., Benet-Buchholz J., Hildebrandt P., Llobet A ., ChemSusChem, 2017,10(3), 551—561
doi: 10.1002/cssc.201601221 URL pmid: 27863077 |
| [19] |
Schorr N. B., Jiang A. G., Rodríguez-López J., Anal. Chem., 2018,90(13), 7848—7854
doi: 10.1021/acs.analchem.8b00730 URL pmid: 29701059 |
| [20] |
Li J. F., Zhang Y. J., Rudnev A. V., Anema J. R., Li S. B., Hong W. J., Rajapandiyan P., Lipkowski J., Wandlowski T., Tian Z. Q., J. Am. Chem. Soc., 2015,137(6), 2400—2408
doi: 10.1021/ja513263j URL pmid: 25625429 |
| [21] |
Wang W., Shao F., Kröger M., Zenobi R., Schlüter A. D., J. Am. Chem. Soc., 2019,141(25), 9867—9871
doi: 10.1021/jacs.9b01765 URL pmid: 31244135 |
| [22] |
Bryce D. A., Kitt J. P., Harris J. M., Anal. Chem., 2018,90(19), 11509—11516
doi: 10.1021/acs.analchem.8b02791 URL pmid: 30175578 |
| [23] |
Zhang C., Li J., Lan L., Cheng J. X., Anal. Chem., 2017,89(8), 4502—4507
doi: 10.1021/acs.analchem.6b04699 URL pmid: 28345862 |
| [24] |
Vogel H., Wright J. K., Jähnig F., Embo J., 1985,4(13), 3625—3631
URL pmid: 3912174 |
| [25] |
Zhang C., Winnard P. T., Dasari S., Kominsky S. L., Doucet M., Jayaraman S., Raman V., Barman I., Chem. Sci., 2018,9(3), 743—753
doi: 10.1039/c7sc02905e URL pmid: 29629144 |
| [26] |
Chen D. D., Xie X. F., Liu J. L., Peng C., J. Chin. Med. Assoc., 2017,80(5), 288—296
doi: 10.1016/j.jcma.2016.11.009 URL pmid: 28325576 |
| [27] |
Batista A. J., Vianna P. G., Ribeiro H. B., Matos C., Gomes A., Sci. Rep. UK, 2019,9(1), 8927—8934
doi: 10.1038/s41598-019-45405-7 URL pmid: 31222126 |
| [28] | Michiels J. E., Vanden B. B., Verstraeten N., Michiels J., Drug Resist. Update., 2016,29(15), 76—89 |
| [29] | Wu W., Feng Y., Tang G., Qiao F., Nally A., Zong Z., Clin. Microbiol. Rev, 2019,32(2), 201—204 |
| [30] | Wu J., Li F., Hu X., Lu J., Sun X., Gao J., Ling D., ACS Central Sci., 2019,5(8), 1366—1376 |
| [31] | Oviaño M., Bou G., Clin. Microbiol. Rev, 2019,32(1), 415—420 |
| [32] |
Bolt F., Cameron J. S., Karancsi T., Simon D., Schaffer R., Rickards T., Hardiman K., Burke A., Bodai Z., Perdones M. A., Rebec M., Balog J., Takats Z., Anal. Chem., 2016,88(19), 9419—9426
doi: 10.1021/acs.analchem.6b01016 URL pmid: 27560299 |
| [33] |
Robert C. B., Thomson M., Vercellone A., Gardner F., Ernst R. K., Larrouy M. G., Nigou J., Biochimie, 2017,141, 16—20
doi: 10.1016/j.biochi.2017.08.008 URL pmid: 28807561 |
| [34] |
Qiu W., Huang Y., Zhao C., Lin Z. S., Lin W. X., Wang Z. J., J. Sci. Food. Agr., 2019,99(9), 4498—4503
doi: 10.1002/jsfa.9695 URL pmid: 30883770 |
| [35] |
Ohlsson P., Evander M., Petersson K., Mellhammar L., Lehmusvuori A., Karhunen U., Soikkeli M., Seppä T., Tuunainen E., Spangar A., Lode P., Rantakokko J. K., Otto G., Scheding S., Soukka T., Wittfooth S., Laurell T., Anal. Chem., 2016,88(19), 9403—9411
doi: 10.1021/acs.analchem.6b00323 URL pmid: 27264110 |
| [36] |
Shu B., Zhang C., Xing D., Biosens. Bioelectron, 2017,97, 360—368
doi: 10.1016/j.bios.2017.06.014 URL pmid: 28624618 |
| [37] |
Zhu Y., Wang Y., Yan Y., Xue H., Environ. Sci. Technol, 2019,53(12), 6895—6905
doi: 10.1021/acs.est.9b01017 URL pmid: 31120737 |
| [38] |
Prudent E., Raoult D., FEMS Microbiol. Rev., 2019,43(1), 88—107
doi: 10.1093/femsre/fuy040 URL pmid: 30418568 |
| [39] |
Lagier J. C., Edouard S., Pagnier I., Mediannikov O., Drancourt M., Raoult D., Clin. Microbiol. Rev., 2015,28(1), 208—236
doi: 10.1128/CMR.00110-14 URL pmid: 25567228 |
| [40] |
Perry J. D., Clin. Microbiol. Rev., 2017,30(2), 449—479
doi: 10.1128/CMR.00097-16 URL pmid: 28122803 |
| [41] |
Tedijanto C., Olesen S. W., Grad Y. H., Lipsitch M., Proc. Natl. Acad. Sci. USA, 2018,115(51), E11988—E11995
doi: 10.1073/pnas.1810840115 URL pmid: 30559213 |
| [42] | Zeiri L., Bronk B. V., Shabtai Y. J., Efrima S., Colloid Surface A, 2002,208(1—3), 357—362 |
| [43] | Defraine V., Fauvart M., Michiels J., Drug Resist . Update., 2018,38(16), 12—26 |
| [44] | Dekter H. E., Orelio C. C., Morsink M. C., Tektas S., Witt R., Leeuwen W. B., Eur. J Clin. Microbiol., 2017,36(1), 81—89 |
| [45] | Oren A., Mana L., Jehlička J., FEMS Microbiol. Lett., 2015,362(6), 22—34 |
| [46] |
Wulf M. W., Willemse E. D., Verduin C. M., Puppels G., Belkum A., Maquelin K., Clin. Microbiol. Infec., 2012,18(2), 147—152
doi: 10.1111/j.1469-0691.2011.03517.x URL pmid: 21854500 |
| [47] |
Choo-Smith L. P., Maquelin K., Vreeswijk T., Appl. Environ. Microb., 2001,67(4), 1461—1469
doi: 10.1128/AEM.67.4.1461-1469.2001 URL pmid: 11282591 |
| [48] | Kunapareddy N., Grun J., Lunsford R., Proc. Spie, 2012,8358, 1—7 |
| [49] | Sarkar N., Sahoo G., Das R., Prusty G., Swain S. K., Eur. J. Pharm. Sci., 2017,109(23), 359—371 |
| [50] | Salesa B., Martí M., Frígols B., Serrano A ., Polymers, 2019,11(3), 453—466 |
| [51] | Bjørnøy S. H., Bassett D. C., Ucar S., Strand B. L., Andreassen J. P., Sikorski P., Acta Biomater., 2016,44(9), 254—266 |
| [52] |
Pahlow S., Meisel S., Cialla M. D., Weber K. R., Popp J., Adv. Drug. Deliv. Rev., 2015,89, 105—120
doi: 10.1016/j.addr.2015.04.006 URL pmid: 25895619 |
| [53] | Kotanen C. N., Martinez. L., Alvarez. R., Simecek J. W., Sensing & Bio-Sensing Research, 2016,8, 20—26 |
| [54] | Silva F. G., Alcaraz E. J., Costa M. M., Oliveira H. P., Mat. Sci. Eng. C-Mater., 2019,99(12), 827—837 |
| [55] |
Lu X., Ge W., Li Q., Wu Y., Jiang H., Wang X., ACS Appl. Mater. Inter, 2014,6(14), 11025—11031
doi: 10.1021/am5016099 URL pmid: 24950258 |
| [56] |
Vergidis P., Moore C. B., Novak F. L., Richardson R., Walker A., Denning D. W., Richardson M. D., Clin. Microbiol. Infec., 2019,5(12), 666—680
doi: 10.1046/j.1469-0691.2001.00350.x URL pmid: 11843907 |
| [57] | Xu Y., Wang H., Luan C., Liu Y., Chen B., Zhao Y., Biosens. Bioelectron, 2018,100(4), 404—410 |
| [58] |
Alamer S., Eissa S., Chinnappan R., Zourob M., Mikrcohim. Acta, 2018,185(3), 164—174
doi: 10.1007/s00604-018-2696-7 URL pmid: 29594804 |
| [59] |
Jung J. Y., Yoon H. K., An S., Lee J. W., Kim Y. J., Park H. C., Lee K., Hwang J. H., Lim S. K., Sci. Rep.-UK., 2018,8(1), 10852—10862
doi: 10.1038/s41598-018-29264-2 URL pmid: 30022122 |
| [60] |
Borer B., Ataman M., Hatzimanikatis V., PLOS Comput. Biol., 2019,15(6), e1007127
doi: 10.1371/journal.pcbi.1007127 URL pmid: 31216273 |
| [61] |
Pande S., Kost C., Trends. Microbiol, 2017,25(5), 349—361
doi: 10.1016/j.tim.2017.02.015 URL pmid: 28389039 |
| [62] | Goo E., Kang Y., Lim J. Y., Ham H., Hwang I., Mbio, 2017,8(1), 7—20 |
| [63] | Driscoll T. P., Verhoeve V. I., Guillotte M. L., Lehman S. S., Rennoll S. A., Beier S. M., Rahman M. S., Azad A. F., Gillespie J. J., Mbio, 2017,8(5), 1456—1466 |
| [64] |
Kelly J., Patrick R., Patrick S., Bell S. E. J., Angew. Chem. Int. Ed., 2018,57(48), 15686—15690
doi: 10.1002/anie.201808185 URL pmid: 30291659 |
| [65] |
Bodelón G., Montes G. V., López P. V., Hill E. H., Hamon C., Sanz O. M. N., Rodal C. S., Costas C., Celiksoy S., Pérez J. I., Scarabelli L., La P. A., Nat. Mater., 2016,15(11), 1203—1211
doi: 10.1038/nmat4720 URL pmid: 27500808 |
| [66] |
Olaniyi O. O., Yang K., Zhu Y. G., Cui L., Appl. Mivrobiol. Biot., 2019,103(3), 1455—1464
doi: 10.1007/s00253-018-9459-6 URL pmid: 30535579 |
| [67] |
Cui L., Yang K., Li H. Z., Zhang H., Su J. Q., Paraskevaidi M., Martin F. L., Ren B., Zhu Y. G., Anal. Chem., 2018,90(8), 5082—5089
doi: 10.1021/acs.analchem.7b05080 URL pmid: 29557648 |
| [68] |
Yang K., Li H. Z., Zhu X., Su J. Q., Ren B., Zhu Y. G., Cui L., Anal. Chem., 2019,91(9), 6296—6303
doi: 10.1021/acs.analchem.9b01064 URL pmid: 30942570 |
| [39] | Xu J., Zhu D., Ibrahim A. D., Allen C. C. R., Gibson C. M., Fowler P. W., Song Y., Huang W. E., Anal. Chem., 2017,89(24), 13305—13312 |
| [70] |
Hong W., Karanja C. W., Abutaleb N. S., Younis W., Zhang X., Seleem M. N., Cheng J. X., Anal. Chem., 2018,90(6), 3737—3743
doi: 10.1021/acs.analchem.7b03382 URL pmid: 29461044 |
| [71] | Xu J. Y., Murphy W. A., Milton D. R., Jimenez C., Rao S. N., Habra M. A., Waguespack S. G., Dadu R., Gagel R. F., Ying A. K., Cabanillas M. E., Weitzman S. P., Busaidy N. L., Sellin R.V., Grubbs E., Sherman S. I., Hu M. I., J. Thorac. Oncol., 2016,101(12), 4871—4877 |
| [72] | Hu P. P., Liu H., Zhen S. J., Li C. M., Huang C. Z., Biosens. Bioelectron., 2015,73(2), 228—233 |
| [73] |
Liu Y., Zhou H., Hu Z., Yu G., Yang D., Zhao J., Biosens. Bioelectron, 2017,94, 131—140
doi: 10.1016/j.bios.2017.02.032 URL pmid: 28262610 |
| [74] |
Tardif S., Pavlenko E., Quazuguel L., Boniface M., Maréchal M., Micha J. S., Gonon L., Mareau V., Gebel G., Bayle G. P., Rieutord F., Lyonnard S., ACS Nano, 2017,11(11), 11306—11316
doi: 10.1021/acsnano.7b05796 URL pmid: 29111665 |
| [75] |
Lin L., Zhang Q., Li X., Qiu M., Jiang X., Jin W., Gu H., Lei D. Y., Ye J., ACS Nano, 2018,12(7), 6492—6503
doi: 10.1021/acsnano.7b08224 URL pmid: 29924592 |
| [76] |
Park J. E., Lee Y., Nam J. M., Nano lett., 2018,18(10), 6475—6482
doi: 10.1021/acs.nanolett.8b02973 URL pmid: 30153413 |
| [77] |
Benzine O., Bruns S., Pan Z., Durst K., Wondraczek L., Adv. Sci, 2018,5(10), 1800916
doi: 10.1002/advs.201800916 URL pmid: 30356973 |
| [78] |
Li J., Yan H., Tan X., Lu Z., Han H., Anal. Chem, 2019,91(6), 3885—3892
doi: 10.1021/acs.analchem.8b04622 URL pmid: 30793591 |
| [79] |
Srivastava S. K., Hamo H. B., Kushmaro A., Marks R. S., Grüner C., Rauschenbach B., Abdulhalim I., Analyst, 2015,140(9), 3201—3209
doi: 10.1039/c5an00209e URL pmid: 25756826 |
| [80] |
Lin Y., Hamme I ., Analyst, 2014,139(15), 3702—3705
doi: 10.1039/c4an00744a URL pmid: 24897935 |
| [81] |
Milazzo L., Gabler T., Pühringer D., Jandova Z., Maresch D., Michlits H., Pfanzagl V., Djinović C. K., Oostenbrink C., Furtmüller P. G., Obinger C., Smulevich G., Hofbauer S., ACS Catal., 2019,9(8), 6766—6782
doi: 10.1021/acscatal.9b00963 URL pmid: 31423350 |
| [82] |
Eryılmaz M., Acar S. E., Çetin D., Boyacı Í. H., Suludere Z., Tamer U., Analyst, 2019,144(11), 3573—3580
doi: 10.1039/c9an00173e URL pmid: 31062777 |
| [83] | Guo L., Ye C., Cui L., Wan K., Chen S., Zhang S., Yu X., Environ. Int, 2019,130(15), 104883—104893 |
| [84] |
Pahlow S., Stöckel S., Pollok S., Cialla-May D., Rösch P., Weber K., Popp J., Anal. Chem, 2016,88(3), 1570—1577
doi: 10.1021/acs.analchem.5b02829 URL pmid: 26705822 |
| [85] | Pang Y., Wan N., Shi L., Wang C., Sun Z., Xiao R., Wang S., Anal. Chim. Acta, 2019,1077(22), 288—296 |
| [86] |
Wang J., Wu X., Wang C., Rong Z., Ding H., Li H., Li S., Shao N., Dong P., Xiao R., Wang S., ACS Appl. Mater. Inter, 2016,8(31), 19958—19967
doi: 10.1021/acsami.6b07528 URL pmid: 27420923 |
| [87] |
Rippa M., Castagna R., Pannico M., Musto P., Borriello G., Paradiso R., Galiero G., Bolletti C. S., Zhou J., Zyss J., Petti L., ACS Sens., 2017,2(7), 947—954
doi: 10.1021/acssensors.7b00195 URL pmid: 28750539 |
| [88] |
Cui L., Zhang D. D., Yang K., Zhang X., Zhu Y. G., Anal. Chem., 2019,91(24), 15345—15354
doi: 10.1021/acs.analchem.9b03996 URL pmid: 31714062 |
| [1] | 陈嘉敏, 曲孝章, 齐国华, 徐蔚青, 金永东, 徐抒平. SERS纳米探针对电刺激过程中细胞内产生活性氧的检测[J]. 高等学校化学学报, 2022, 43(6): 20220033. |
| [2] | 王园月, 安梭梭, 郑旭明, 赵彦英. 5-巯基-1, 3, 4-噻二唑-2-硫酮微溶剂团簇的光谱和理论计算研究[J]. 高等学校化学学报, 2022, 43(10): 20220354. |
| [3] | 薛谨, 曹小卫, 刘依帆, 王敏. 纸质空心金纳米笼SERS传感器的制备及对非小细胞肺癌患者痰液中miRNAs的快速高灵敏检测[J]. 高等学校化学学报, 2021, 42(8): 2393. |
| [4] | 陈峰, 程娜, 赵健伟, 宋易恬, 孙燕燕, 娄鑫梨, 童夏燕. 纳米颗粒银层的电沉积机理及SERS效应[J]. 高等学校化学学报, 2021, 42(6): 1891. |
| [5] | 潘菁, 徐敏敏, 袁亚仙, 姚建林. 基于表面增强拉曼光谱快速检测纺织品禁用染料[J]. 高等学校化学学报, 2021, 42(12): 3716. |
| [6] | 张晓荣, 陈岚岚, 胡善文. 基于分子识别的细菌检测研究进展[J]. 高等学校化学学报, 2021, 42(11): 3468. |
| [7] | 王瑞雪, 尹冬梅, 宋永新, 单桂晔. CuS/Ag2S纳米复合物的制备及类过氧化物酶性质[J]. 高等学校化学学报, 2020, 41(6): 1218. |
| [8] | 张辉, 张晨杰, 徐敏敏, 袁亚仙, 姚建林. SERS-HPLC联用技术对邻氨基苯硫酚与邻碘苯甲酰氯反应的监测[J]. 高等学校化学学报, 2020, 41(11): 2496. |
| [9] | 魏曼曼,路皓翔,杨辉华. 基于RF_AdaBoost模型的血液种属鉴别算法[J]. 高等学校化学学报, 2020, 41(1): 94. |
| [10] | 魏馨, 邓要亮, 郑旭明, 赵彦英. 2-氨基苯并噻唑的结构及激发态质子转移动力学[J]. 高等学校化学学报, 2019, 40(8): 1679. |
| [11] | 曹培明, 尤静林, 周灿栋, LU Liming, 王建, 王敏, 丁雅妮, 徐琰东. CaF2-Al2O3-MgO电渣微结构的原位拉曼光谱及27Al魔角旋转核磁共振研究[J]. 高等学校化学学报, 2019, 40(6): 1242. |
| [12] | 刘秋娜, 许文文, 刘茂祝, 王惠钢, 郑旭明. 高分辨偏正拉曼光谱对丙酸酐C=O振动模的拉曼光谱非一致效应研究[J]. 高等学校化学学报, 2019, 40(5): 932. |
| [13] | 徐琰东,尤静林,王建,龚晓晔,丁雅妮,曹培明,郑少波,吴永全,余仲达. Bi4B2O9晶体及其熔体结构的高温原位拉曼光谱研究[J]. 高等学校化学学报, 2019, 40(10): 2143. |
| [14] | 韩东来,李博珣,杨硕,闫永胜,杨丽丽,刘洋,李春香. Fe3O4@Au核-壳纳米复合材料的制备及对农药福美双的SERS检测研究[J]. 高等学校化学学报, 2019, 40(10): 2067. |
| [15] | 冯微, 王博蔚, 郑艳, 姜洋. 金纳米粒子簇的制备及表面增强拉曼光谱[J]. 高等学校化学学报, 2018, 39(9): 1875. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||