

Chem. J. Chinese Universities ›› 2019, Vol. 40 ›› Issue (12): 2465.doi: 10.7503/cjcu20190450
• Analytical Chemistry • Previous Articles Next Articles
Zhiqing ZHANG( ),Shanshan WANG,Zichen ZHANG,Jie MA,Xiufeng WANG(
),Shanshan WANG,Zichen ZHANG,Jie MA,Xiufeng WANG( ),Ting ZHOU,Fang WANG,Guodong ZHANG
),Ting ZHOU,Fang WANG,Guodong ZHANG
Received:2019-08-12
Online:2019-12-04
Published:2019-12-04
Contact:
Zhiqing ZHANG,Xiufeng WANG
E-mail:zhangzq@upc.edu.cn;xfwang@upc.edu.cn
Supported by:CLC Number:
TrendMD:
Zhiqing ZHANG,Shanshan WANG,Zichen ZHANG,Jie MA,Xiufeng WANG,Ting ZHOU,Fang WANG,Guodong ZHANG. Rolling Circle Amplification-based Polyvalent Molecular Beacon Probe for Signal-amplifying and Sensitive-Detection of Thrombin †[J]. Chem. J. Chinese Universities, 2019, 40(12): 2465.
| Name | DNA sequence(5'-3') | Source |
|---|---|---|
| Circle DNA1 | TGTCTTCGCCTTCTTGTTTCCTTTCCTTGAAACTTCTTCCTTTCT- | TaKaRa Biotechnology(Dalian) Co., Ltd. |
| TTCTTTCGACTAAGCACC | ||
| Primer DNA2 | GGCGAAGACAGGTGCTTAGTC | TaKaRa Biotechnology(Dalian) Co., Ltd. |
| Repeated sequence of | GGTGCTTAGTCGAAAGAAAGAAAGGAAGAAGTTTCAAGGAA- | TaKaRa Biotechnology(Dalian) Co., Ltd. |
| RCA product | AGGAAACAAGAAGGCGAAGACA | |
| MB | (FAM)GGAAGAAGTGGTTGGTGTGGTTGGCCCCTGTCTTCGC- | Sangon Biotechnology(Shanghai) Co., Ltd. |
| CTTCTTGTTTCCTTTCCTTGAAACTTCTTCC(DABCYL) | ||
| FC short chain | TTTCTTTCTTTCGACTAAGCACC | Sangon Biotechnology(Shanghai) Co., Ltd. |
| 63-cDNA | GGTGCTTAGTCGAAAGAAAGAAAGGAAGAAGTTTCAAGGAA- | Sangon Biotechnology(Shanghai) Co., Ltd. |
| AGGAAACAAGAAGGCGAAGACA |
| Name | DNA sequence(5'-3') | Source |
|---|---|---|
| Circle DNA1 | TGTCTTCGCCTTCTTGTTTCCTTTCCTTGAAACTTCTTCCTTTCT- | TaKaRa Biotechnology(Dalian) Co., Ltd. |
| TTCTTTCGACTAAGCACC | ||
| Primer DNA2 | GGCGAAGACAGGTGCTTAGTC | TaKaRa Biotechnology(Dalian) Co., Ltd. |
| Repeated sequence of | GGTGCTTAGTCGAAAGAAAGAAAGGAAGAAGTTTCAAGGAA- | TaKaRa Biotechnology(Dalian) Co., Ltd. |
| RCA product | AGGAAACAAGAAGGCGAAGACA | |
| MB | (FAM)GGAAGAAGTGGTTGGTGTGGTTGGCCCCTGTCTTCGC- | Sangon Biotechnology(Shanghai) Co., Ltd. |
| CTTCTTGTTTCCTTTCCTTGAAACTTCTTCC(DABCYL) | ||
| FC short chain | TTTCTTTCTTTCGACTAAGCACC | Sangon Biotechnology(Shanghai) Co., Ltd. |
| 63-cDNA | GGTGCTTAGTCGAAAGAAAGAAAGGAAGAAGTTTCAAGGAA- | Sangon Biotechnology(Shanghai) Co., Ltd. |
| AGGAAACAAGAAGGCGAAGACA |
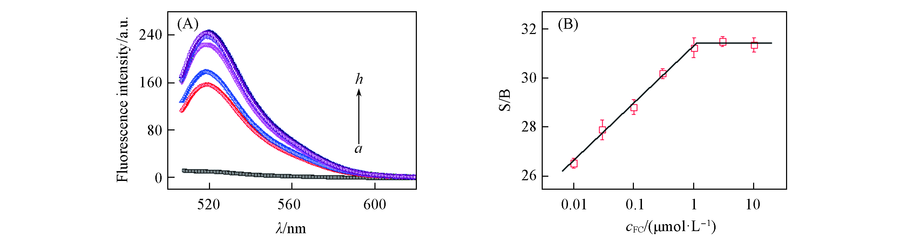
Fig.3 Fluorescence spectra(A) of different concentrations of FC and the signal-to-background ratio(S/B)(B) cMB=30 nmol/L. a. control; cFC:cRCA:cMB : b. 0:1:3; c. 1:1:3; d.10:1:3; e. 30:1:3; f. 100:1:3; g. 300:1:3; h.1000:1:3.
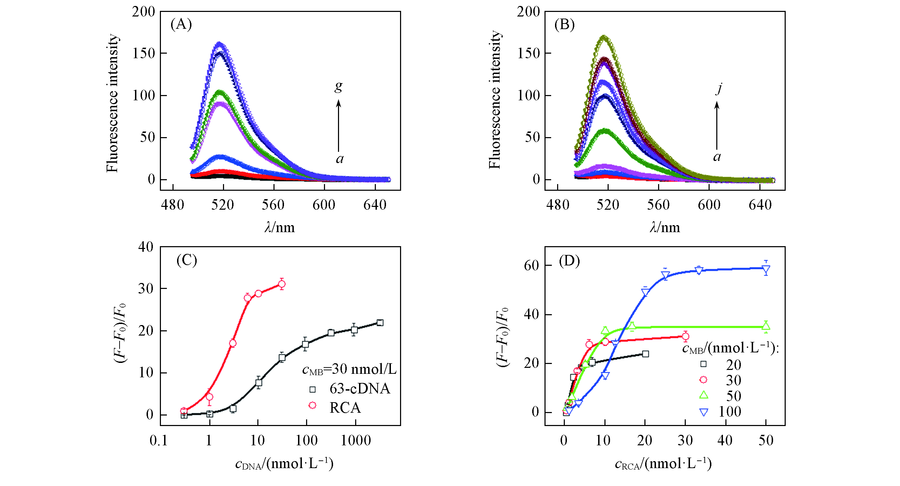
Fig.4 Fluorescence spectra(A, B) and relative fluorescence intensity ratios(C, D) under different conditions (A) a. control; cRCA:cMB : b. 1:100; c.1:30; d.1:10; e.1:5; f. 1:3; g. 1:1. cMB=30 nmol/L. (B) a. Control; c63-cDNA:cMB: b. 1:100; c.1:30; d.1:10; e.1:3; f. 1:1; g. 3:1; h. 10:1; i. 30:1; j. 100:1. cMB=30 nmol/L. (C) RCA-MB assembly and 63-cDNA-MB assembly with 30 nmol/L MB; (D) RCA-MB assembly with different concentrations of MB.
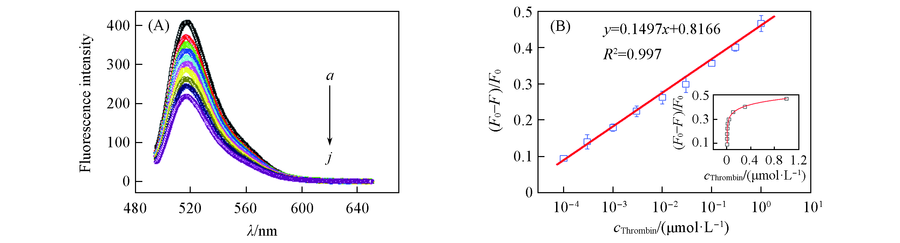
Fig.5 Detection sensitivity of MB-RCA for thrombin (A) Fluorescence spectra. cMB =100 nmol/L. cThrombin/(μmol·L-1): a. 0; b. 0.0001; c. 0.0003; d. 0.001; e. 0.003;f. 0.01; g. 0.03; h. 0.1; i. 0.3; j. 1. (B) Relative fluorescence intensity ratio vs. the concentration of thrombin.
| Method | Linear range/(nmol·L-1) | Detection limit/(nmol·L-1) | Ref. |
|---|---|---|---|
| Electrochemical sensor based on aptamers | 40—100 | 10 | [ |
| Aptamer biosensor based on guanine-quenching | | 0.81 | [ |
| Fluorescence detection based on molecular beacon | 0—30 | 0.83 | [ |
| Fluorescence detection based on dual quenching molecular beacon | 0.4—40 | 0.18 | [ |
| Polyvalent molecular beacon probe base on RCA | 0.1—30 | 0.20 | This work |
| Method | Linear range/(nmol·L-1) | Detection limit/(nmol·L-1) | Ref. |
|---|---|---|---|
| Electrochemical sensor based on aptamers | 40—100 | 10 | [ |
| Aptamer biosensor based on guanine-quenching | | 0.81 | [ |
| Fluorescence detection based on molecular beacon | 0—30 | 0.83 | [ |
| Fluorescence detection based on dual quenching molecular beacon | 0.4—40 | 0.18 | [ |
| Polyvalent molecular beacon probe base on RCA | 0.1—30 | 0.20 | This work |
| [1] |
Arsenault K. A., Hirsh J., Whitlock R. P., Eikelboom J. W ., Nat. Rev. Cardiol., 2012,9(7), 402— 414
doi: 10.1038/nrcardio.2012.61 URL |
| [2] |
Petrlova J., Hansen F. C., van der Plas M. J. A., Huber R. G., MöRgelin M., Malmsten M ., Proc. Natl. Acad. Sci. USA, 2017,114(21), E4213— E4222
doi: 10.1073/pnas.1619609114 URL pmid: 28473418 |
| [3] |
Lin K. Y., Kwong G. A., Warren A. D., Wood D. K., Bhatia S. N ., ACS Nano, 2013,7(10), 9001— 9009
doi: 10.1021/nn403550c URL pmid: 24015809 |
| [4] |
Pavlov V., Shlyahovsky B., Willner I ., J. Am. Chem. Soc., 2005,127(18), 6522— 6523
doi: 10.1021/ja050678k URL pmid: 15869259 |
| [5] | Du S. S., Li Y., Guo L., Li P. Y., Chai Z. L., Wang T., Quan D. Q., He J. L ., Chem. J. Chinese Universities, 2018,39(11), 2445— 2450 |
| ( 杜闪闪, 李阳, 郭磊, 李鹏羽, 柴智龙, 王涛, 全东琴, 何军林 . 高等学校化学学报, 2018,39(11), 2445— 2450) | |
| [6] |
Kim Y., Cao Z. H., Tan W. H ., Proc. Natl. Acad. Sci. USA, 2008,105(15), 5664— 5669
doi: 10.1073/pnas.0711803105 URL pmid: 18398007 |
| [7] |
Riccardi C., Russo Krauss I., Musumeci D., Morvan F., Meyer A., Vasseur J. J ., ACS Appl. Mater. Interfaces, 2017,9(41), 35574— 35587
doi: 10.1021/acsami.7b11195 URL pmid: 28849915 |
| [8] |
Feagin T. A., Maganzini N., Soh H. T ., ACS Sensors, 2018,3(9), 1611— 1615
doi: 10.1021/acssensors.8b00516 URL pmid: 30156834 |
| [9] |
Heckel A., Mayer Günter ., J. Am. Chem. Soc., 2005,127(3), 822— 823
doi: 10.1021/ja043285e URL pmid: 15656605 |
| [10] |
Tang Z. W., Mallikaratchy P., Yang R. H., Kim Y., Zhu Z., Wang H., Tan W. H ., J. Am. Chem. Soc., 2008,130(34), 11268— 11269
doi: 10.1021/ja804119s URL pmid: 18680291 |
| [11] |
Bai Y. L., Li Y. P, Zhang D. P., Wang H. L., Zhao Q ., Anal. Chem., 2017,89(17), 9467— 9473
doi: 10.1021/acs.analchem.7b02313 URL pmid: 28763192 |
| [12] |
Fu Q. Q., Wu Z., Du D., Zhu C. Z., Lin Y. H., Tang Y ., ACS Sensors, 2017,2(6), 789— 795
doi: 10.1021/acssensors.7b00156 URL pmid: 28723117 |
| [13] |
Chen Z. B., Tan L. L., Hu L. Y., Zhang Y. M., Wang S. X., Lv F. Y ., ACS Appl. Mater. Interfaces, 2016,8(1), 102— 108
doi: 10.1021/acsami.5b08975 URL pmid: 26558607 |
| [14] |
Shen G., Zhang H., Yang C. R., Yang Q. F., Tang Y. L ., Anal. Chem., 2016,89(1), 548— 551
doi: 10.1021/acs.analchem.6b04247 URL pmid: 27958723 |
| [15] | Liu S. Z., Zhang Z. Q., Wang F., Zhou T., Wang X. F., Zhang G. D., Liu T. T., Zhang H. Z ., Acta Phys.-Chim. Sinica, 2017,33(10), 2052— 2057 |
| ( 刘淑贞, 张志庆, 王芳, 周亭, 王秀凤, 张国栋, 刘婷婷, 张洪芝 . 物理化学学报, 2017,33(10), 2052— 2057) | |
| [16] | Zhang H. Z., Zhang Z. Q., Wang F., Zhou T., Wang X. F., Zhang G. D., Liu T. T., Liu S. Z ., Acta Phys.-Chim. Sinica, 2017,33(8), 1520— 1532 |
| ( 张洪芝, 张志庆, 王芳, 周亭, 王秀凤, 张国栋, 刘婷婷, 刘淑贞 . 物理化学学报, 2017,33(8), 1520— 1532) | |
| [17] |
Zhang Z. Q., Zhang H. Z., Wang F., Zhang G. D., Zhou T., Wang X. F., Liu S. Z., Liu T. T ., Macromol. Rapid Commun., 2018,39(15), 1800263
doi: 10.1002/marc.201800263 URL pmid: 29952041 |
| [18] |
Zhang Z. Q., Liu S. Z, Zhou T., Zhang H. Z, Wang F., Zhang G. D., Wang X. F., Liu T. T ., Langmuir, 2018,34(30), 8904— 8909
doi: 10.1021/acs.langmuir.8b01505 URL pmid: 29945443 |
| [19] |
Zhang Z. Q., Eckert M. A., Ali M. M., Liu L. N., Kang D. K., Chang E., Sender L. S., Fruman D. A., Zhao W. A ., ChemBioChem, 2014,15(9), 1268— 1273
doi: 10.1002/cbic.201402100 URL |
| [20] |
Zhang Z. Q., Ali M. M., Eckert M. A., Kang D. K., Chen Y. Y., Sender L. S., Fruman D. A., Zhao W. A ., Biomaterials, 2013,34(37), 9728— 9735
doi: 10.1016/j.biomaterials.2013.08.079 URL |
| [21] |
Ali M. M., Li F., Zhang Z. Q., Zhang K. X., Kang D. K., Ankrum J. A., Le X. C., Zhao W. A ., Chem. Soc. Rev., 2014,43(10), 3324— 3341
doi: 10.1039/c3cs60439j URL pmid: 24643375 |
| [22] | Wen Y. Q., Long Q., Zhang Y. Y., Li H. T ., Journal of Analytical Sciences, 2017,33(2), 177— 182 |
| ( 文艳清, 龙倩, 张友玉, 李海涛 . 分析科学学报, 2017,33(2), 177— 182) | |
| [23] | Zhong Z. H., Yang X. H., Wang K. M., Tan W. H., Li H. M., Meng X. X., Guo Q. P ., Life. Sci. Res., 2006,10(3), 206— 209 |
| ( 仲志鸿, 杨小海, 王柯敏, 谭蔚泓, 李惠敏, 孟祥贤, 郭秋平 . 生命科学研究, 2006,10(3), 206— 209) | |
| [24] |
Zhang L. B., Zhu J. B., Li T., Wang E ., Anal. Chem., 2011,83(23), 8871— 8876
doi: 10.1021/ac2006763 URL pmid: 22017597 |
| [25] |
Zhao Y. X., Chen F., Li Q., Wang L. H., Fan C. H ., Chem. Rev., 2015,115(22), 12491— 12545
doi: 10.1021/acs.chemrev.5b00428 URL pmid: 26551336 |
| [26] |
Ikebukuro K., Kiyohara C., Sode K ., Biosens. Bioelectron., 2005,20(10), 2168— 2172
doi: 10.1016/j.bios.2004.09.002 URL pmid: 15741093 |
| [27] |
Wang W. J., Chen C. L., Qian M. X., Zhao X. S ., Sens. Actuators B, 2008,129(1), 211— 217
doi: 10.1016/j.snb.2007.07.125 URL |
| [28] | Zhai K., Li F. Q., Shi B. A., Xiang D. S ., Chinese J. Anal. Chem., 2017,45(10), 1462— 1466 |
| ( 翟琨, 李奉权, 史伯安, 向东山 . 分析化学, 2017,45(10), 1462— 1466) |
| [1] | XIE Chen, CHEN Na, YANG Yanbing, YUAN Quan. Recent Progress of Aptamer Functionalized Two-dimensional Materials Field Effect Transistor Sensors [J]. Chem. J. Chinese Universities, 2021, 42(11): 3406. |
| [2] | ZHAO Zhuo, WANG Xueqiang. Investigations upon the Bioconjugation-based Construction Technologies and Applications of Aptamer-drug Conjugates [J]. Chem. J. Chinese Universities, 2021, 42(11): 3367. |
| [3] | LIU Xuejiao, YANG Fan, LIU Shuang, ZHANG Chunjuan, LIU Qiaoling. Progress in Aptamer-targeted Membrane Protein Recognition and Functional Regulation [J]. Chem. J. Chinese Universities, 2021, 42(11): 3277. |
| [4] | HUANG Ling, ZHUANG Zijian, LI Xiang, SHI Muling, LIU Gaoqiang. Advances in Molecular Recognition of Exosomes Based on Aptamers [J]. Chem. J. Chinese Universities, 2021, 42(11): 3493. |
| [5] | LIU Ke, JIN Yu, LIANG Jiangong, WU Yuan. Research Progress on Improving the Binding Affinity of Aptamers through Chemical Modification [J]. Chem. J. Chinese Universities, 2021, 42(11): 3477. |
| [6] | ZHANG Xiaorong, CHEN Lanlan, HU Shanwen. Advances in Bacteria Biosensing Based on Molecular Recognition [J]. Chem. J. Chinese Universities, 2021, 42(11): 3468. |
| [7] | JI Cailing, CHENG Xing, TAN Jie, YUAN Quan. Selection of Functionalized Aptamers and Their Applications in Molecular Recognition [J]. Chem. J. Chinese Universities, 2021, 42(11): 3457. |
| [8] | REN Yushuang, GUO Yuanyuan, LIU Xueyi, SONG Jie, ZHANG Chuan. Platinum(Ⅳ) Prodrug-grafted Phosphorothioate DNA and Its Self-assembled Nanostructure for Targeted Drug Delivery [J]. Chem. J. Chinese Universities, 2020, 41(8): 1721. |
| [9] | DU Xianchao, HAO Hongxia, QIN Anjun, TANG Benzhong. Detection of Cocaine Based on the System of AIEgen, Aptamer and Exonuclease Ⅰ † [J]. Chem. J. Chinese Universities, 2020, 41(3): 411. |
| [10] | DONG Qian, LI Zhaoqian, PENG Tianhuan, CHEN Zhuo, TAN Weihong. Progress on Aptamer for Cancer Theranostics [J]. Chem. J. Chinese Universities, 2020, 41(12): 2648. |
| [11] |
LI Delei,GU Mengqiao,WANG Min,CHI Kuanneng,ZHANG Xi,DENG Yan,MA Yuchan,HU Rong,YANG Yunhui.
Preparation of Thrombin Aptasensor Based on the Metal-organic Framework Fe-MIL-88N |
| [12] | WANG Chunyan,JIANG Xiaoqing,ZHOU Bo. An Electrochemical Biosensor Based on Cu-TPA for Determination of Aflatoxin B1 † [J]. Chem. J. Chinese Universities, 2019, 40(11): 2301. |
| [13] | LIU Zhongcheng, LIU Shifang, ZHANG Su, YANG Yanlei, LI Fei, ZHANG Nan, YUAN Xin, ZHANG Yanfen. Structure Prediction and Screening of Oligonucleotide Aptamers Target Cε3-Cε4 Protein† [J]. Chem. J. Chinese Universities, 2019, 40(1): 83. |
| [14] | DU Shanshan, LI Yang, GUO Lei, LI Pengyu, CHAI Zhilong, WANG Tao, QUAN Dongqin, HE Junlin. Modification of Aptamer TBA with Extra Functional Groups and the Biological Activities† [J]. Chem. J. Chinese Universities, 2018, 39(11): 2445. |
| [15] | CHEN Dongxing, SHI Jinyu, CHEN Qiufang, ZHANG Rui, GONG Guoqing, XU Yungen, ZHU Qihua. Design, Synthesis and Biological Evaluation of Thrombin Inhibitors with 1,2,3,4-Tetrahydrobenzo[4,5]imidazo[1,2-a]pyrazine Nucleus† [J]. Chem. J. Chinese Universities, 2017, 38(6): 1059. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||