

Chem. J. Chinese Universities ›› 2019, Vol. 40 ›› Issue (2): 288.doi: 10.7503/cjcu20180653
• Physical Chemistry • Previous Articles Next Articles
XU Yan, LIU Cui*( ), HAN Chengjuan, PAN Mingyu, SUN Zhaoqi, HAN Bingyu, YANG Zhongzhi
), HAN Chengjuan, PAN Mingyu, SUN Zhaoqi, HAN Bingyu, YANG Zhongzhi
Received:2018-09-26
Online:2019-02-10
Published:2018-11-19
Contact:
LIU Cui
E-mail:liuc@lnnu.edu.cn
Supported by:CLC Number:
TrendMD:
XU Yan,LIU Cui,HAN Chengjuan,PAN Mingyu,SUN Zhaoqi,HAN Bingyu,YANG Zhongzhi. Development of Polarization Force Field for Guanine and Amino Acid Residues Systems†[J]. Chem. J. Chinese Universities, 2019, 40(2): 288.
| Hydrogen bond | k' | C | rlp,H | D |
|---|---|---|---|---|
| lpN307102, H-N101106 | 0.7010 | 0.0931 | 1.3019 | 0.0319 |
| lpN307102, H-N101109 | 0.5438 | 0.0931 | 1.3019 | 0.0319 |
| lpN307102, H-N101111 | 0.5840 | 0.0931 | 1.3019 | 0.0319 |
| lpN307202, H-N101111 | 0.5744 | 0.2500 | 2.0000 | 0.0500 |
| lpO308102, H-N101116 | 0.5800 | 0.0931 | 1.5019 | 0.0319 |
| lpO308102, H-N101117 | 0.5450 | 0.0931 | 1.3019 | 0.0319 |
| lpO308201, H-N101106 | 0.6000 | 0.0792 | 2.0874 | 0.3202 |
| lpO308201, H-N101116 | 0.5850 | 0.0506 | 1.2523 | 0.0536 |
| lpO308201, H-N101117 | 0.6030 | 0.0931 | 1.3019 | 0.0319 |
| lpO308207, H-N101106 | 0.4800 | 0.0931 | 1.3019 | 0.0319 |
| lpO308207, H-N101111 | 0.6570 | 0.0951 | 1.5019 | 0.0519 |
| lpS316102, H-N101116 | 0.8000 | 0.0931 | 1.3019 | 0.0319 |
| lpS316102, H-N101117 | 0.6400 | 0.0931 | 1.3019 | 0.0319 |
Table 1 Parameters k', C, rlp,H and D in the function kHB(Rlp,H)
| Hydrogen bond | k' | C | rlp,H | D |
|---|---|---|---|---|
| lpN307102, H-N101106 | 0.7010 | 0.0931 | 1.3019 | 0.0319 |
| lpN307102, H-N101109 | 0.5438 | 0.0931 | 1.3019 | 0.0319 |
| lpN307102, H-N101111 | 0.5840 | 0.0931 | 1.3019 | 0.0319 |
| lpN307202, H-N101111 | 0.5744 | 0.2500 | 2.0000 | 0.0500 |
| lpO308102, H-N101116 | 0.5800 | 0.0931 | 1.5019 | 0.0319 |
| lpO308102, H-N101117 | 0.5450 | 0.0931 | 1.3019 | 0.0319 |
| lpO308201, H-N101106 | 0.6000 | 0.0792 | 2.0874 | 0.3202 |
| lpO308201, H-N101116 | 0.5850 | 0.0506 | 1.2523 | 0.0536 |
| lpO308201, H-N101117 | 0.6030 | 0.0931 | 1.3019 | 0.0319 |
| lpO308207, H-N101106 | 0.4800 | 0.0931 | 1.3019 | 0.0319 |
| lpO308207, H-N101111 | 0.6570 | 0.0951 | 1.5019 | 0.0519 |
| lpS316102, H-N101116 | 0.8000 | 0.0931 | 1.3019 | 0.0319 |
| lpS316102, H-N101117 | 0.6400 | 0.0931 | 1.3019 | 0.0319 |
| Class | V2/(kJ·mol-1) | Class | V2/(kJ·mol-1) | Class | V2/(kJ·mol-1) |
|---|---|---|---|---|---|
| 84-71-75-75 | 6.90 | 83-71-79-81 | 5.65 | 75-73-76-71 | 41.84 |
| 84-71-75-72 | 6.90 | 78-72-75-75 | 17.36 | 75-73-76-82 | 41.84 |
| 84-71-76-73 | 7.11 | 75-72-78-71 | 20.08 | 83-74-78-71 | 10.04 |
| 84-71-76-82 | 7.11 | 75-72-78-74 | 20.08 | 83-74-78-72 | 10.04 |
| 78-71-79-75 | 5.65 | 76-73-75-75 | 10.67 | 71-75-75-73 | 22.80 |
| 83-71-79-71 | 5.65 | 76-73-75-79 | 6.28 | 72-75-75-73 | 22.80 |
Table 2 Dihedral angle parameters
| Class | V2/(kJ·mol-1) | Class | V2/(kJ·mol-1) | Class | V2/(kJ·mol-1) |
|---|---|---|---|---|---|
| 84-71-75-75 | 6.90 | 83-71-79-81 | 5.65 | 75-73-76-71 | 41.84 |
| 84-71-75-72 | 6.90 | 78-72-75-75 | 17.36 | 75-73-76-82 | 41.84 |
| 84-71-76-73 | 7.11 | 75-72-78-71 | 20.08 | 83-74-78-71 | 10.04 |
| 84-71-76-82 | 7.11 | 75-72-78-74 | 20.08 | 83-74-78-72 | 10.04 |
| 78-71-79-75 | 5.65 | 76-73-75-75 | 10.67 | 71-75-75-73 | 22.80 |
| 83-71-79-71 | 5.65 | 76-73-75-79 | 6.28 | 72-75-75-73 | 22.80 |
| Class | ν/(kJ·mol-1) | Class | ν/(kJ·mol-1) | Class | ν/(kJ·mol-1) | Class | ν/(kJ·mol-1) |
|---|---|---|---|---|---|---|---|
| 72-75-75-71 | 41.84 | 71-72-79-81 | 41.84 | 79-78-71-83 | 41.84 | 75-75-71-84 | 41.84 |
| 79-75-75-71 | 41.84 | 71-75-79-81 | 41.84 | 79-79-71-83 | 41.84 | 76-75-71-84 | 41.84 |
| 79-75-75-73 | 41.84 | 73-71-76-82 | 41.84 | 78-83-74-83 | 87.86 | 79-75-71-84 | 41.84 |
| 71-71-79-81 | 41.84 | 79-75-71-83 | 41.84 |
Table 3 Improper dihedral angle parameters
| Class | ν/(kJ·mol-1) | Class | ν/(kJ·mol-1) | Class | ν/(kJ·mol-1) | Class | ν/(kJ·mol-1) |
|---|---|---|---|---|---|---|---|
| 72-75-75-71 | 41.84 | 71-72-79-81 | 41.84 | 79-78-71-83 | 41.84 | 75-75-71-84 | 41.84 |
| 79-75-75-71 | 41.84 | 71-75-79-81 | 41.84 | 79-79-71-83 | 41.84 | 76-75-71-84 | 41.84 |
| 79-75-75-73 | 41.84 | 73-71-76-82 | 41.84 | 78-83-74-83 | 87.86 | 79-75-71-84 | 41.84 |
| 71-71-79-81 | 41.84 | 79-75-71-83 | 41.84 |
| X—H…Y | Sum | LX—Y/nm | LH—Y/nm | θ/(°) | X—H…Y | Sum | LX—Y/nm | LH—Y/nm | θ/(°) |
|---|---|---|---|---|---|---|---|---|---|
| O…HN | 13 | 0.28 | 0.19 | 158.9 | S…HN | 2 | 0.34 | 0.25 | 154.8 |
| N…HN | 13 | 0.29 | 0.21 | 150.2 | O…HS | 1 | 0.34 | 0.24 | 123.2 |
| O…HO | 1 | 0.27 | 0.19 | 142.8 | N…HS | 1 | 0.34 | 0.23 | 131.7 |
| N…HO | 1 | 0.28 | 0.20 | 144.4 |
Table 4 Average hydrogen bond length(L) and hydrogen bond angle(θ) of complexes
| X—H…Y | Sum | LX—Y/nm | LH—Y/nm | θ/(°) | X—H…Y | Sum | LX—Y/nm | LH—Y/nm | θ/(°) |
|---|---|---|---|---|---|---|---|---|---|
| O…HN | 13 | 0.28 | 0.19 | 158.9 | S…HN | 2 | 0.34 | 0.25 | 154.8 |
| N…HN | 13 | 0.29 | 0.21 | 150.2 | O…HS | 1 | 0.34 | 0.24 | 123.2 |
| O…HO | 1 | 0.27 | 0.19 | 142.8 | N…HS | 1 | 0.34 | 0.23 | 131.7 |
| N…HO | 1 | 0.28 | 0.20 | 144.4 |
| Complex | Charge/e | ||||||
|---|---|---|---|---|---|---|---|
| N6 | O9 | N10 | H11 | N13 | H15 | N16 | |
| G3Arg1 | -0.046* | 0.012 | 0.012 | 0.018 | -0.045* | 0.022 | 0.013 |
| G3Arg2 | -0.048* | 0.012 | 0.012 | 0.020 | -0.046* | 0.021 | 0.014 |
| G4Arg1 | 0.017 | 0.008 | 0.009 | -0.057* | 0.041 | 0.019 | -0.015 |
| G4Arg2 | 0.017 | 0.008 | 0.009 | -0.057* | 0.042 | 0.019 | -0.015 |
| G5Arg1 | -0.037 | 0.014 | 0.012 | 0.019 | -0.072* | 0.024 | 0.014 |
| G5Arg2 | -0.038 | 0.013 | 0.012 | 0.019 | -0.071* | 0.024 | 0.014 |
| G6Arg1 | -0.070* | 0.008 | 0.011 | 0.016 | -0.050 | 0.016 | 0.014 |
| G6Arg2 | -0.070* | 0.008 | 0.012 | 0.016 | -0.046 | 0.016 | 0.013 |
| G3Lys | -0.060* | 0.017 | 0.013 | 0.023 | -0.055* | 0.028 | 0.021 |
| G1Asn1 | 0.002 | 0.001 | -0.011* | -0.021* | 0.002 | 0.001 | 0.034* |
| G2Asn1 | 0 | -0.018* | 0.003 | -0.003 | -0.036* | 0.041* | -0.010 |
| G3Asn2 | -0.015* | 0.003 | 0.012 | 0.008 | -0.001* | 0.007 | -0.008 |
| G1Ser | 0.002 | 0.002 | -0.003* | -0.017* | 0.005 | 0.004 | 0.019* |
| G2Ser | 0.002 | -0.011* | 0.011 | 0.006 | -0.025* | 0.033* | -0.010 |
| G1Cys | 0.001 | 0.001 | 0.006* | -0.002* | 0.004 | 0.003 | -0.004 |
| G2Cys | 0.001 | -0.004* | 0.011 | 0.005 | -0.012* | 0.009* | -0.010 |
Table 5 Change of atomic charges in isolated base G and hydrogen bonded complexes
| Complex | Charge/e | ||||||
|---|---|---|---|---|---|---|---|
| N6 | O9 | N10 | H11 | N13 | H15 | N16 | |
| G3Arg1 | -0.046* | 0.012 | 0.012 | 0.018 | -0.045* | 0.022 | 0.013 |
| G3Arg2 | -0.048* | 0.012 | 0.012 | 0.020 | -0.046* | 0.021 | 0.014 |
| G4Arg1 | 0.017 | 0.008 | 0.009 | -0.057* | 0.041 | 0.019 | -0.015 |
| G4Arg2 | 0.017 | 0.008 | 0.009 | -0.057* | 0.042 | 0.019 | -0.015 |
| G5Arg1 | -0.037 | 0.014 | 0.012 | 0.019 | -0.072* | 0.024 | 0.014 |
| G5Arg2 | -0.038 | 0.013 | 0.012 | 0.019 | -0.071* | 0.024 | 0.014 |
| G6Arg1 | -0.070* | 0.008 | 0.011 | 0.016 | -0.050 | 0.016 | 0.014 |
| G6Arg2 | -0.070* | 0.008 | 0.012 | 0.016 | -0.046 | 0.016 | 0.013 |
| G3Lys | -0.060* | 0.017 | 0.013 | 0.023 | -0.055* | 0.028 | 0.021 |
| G1Asn1 | 0.002 | 0.001 | -0.011* | -0.021* | 0.002 | 0.001 | 0.034* |
| G2Asn1 | 0 | -0.018* | 0.003 | -0.003 | -0.036* | 0.041* | -0.010 |
| G3Asn2 | -0.015* | 0.003 | 0.012 | 0.008 | -0.001* | 0.007 | -0.008 |
| G1Ser | 0.002 | 0.002 | -0.003* | -0.017* | 0.005 | 0.004 | 0.019* |
| G2Ser | 0.002 | -0.011* | 0.011 | 0.006 | -0.025* | 0.033* | -0.010 |
| G1Cys | 0.001 | 0.001 | 0.006* | -0.002* | 0.004 | 0.003 | -0.004 |
| G2Cys | 0.001 | -0.004* | 0.011 | 0.005 | -0.012* | 0.009* | -0.010 |
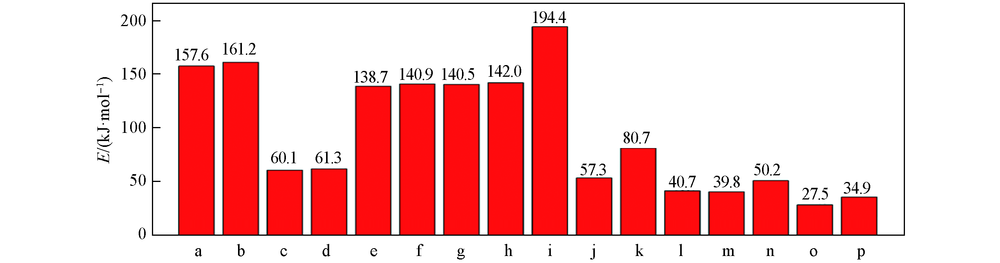
Fig.5 Hydrogen bond energies of hydrogen bond complexesa. G3Arg1; b. G3Arg2; c. G4Arg1; d. G4Arg2; e. G5Arg1; f. G5Arg2; g. G6Arg1; h. G6Arg2; i. G3Lys; j. G1Asn1; k. G2Asn1; l. G3Asn2; m. G1Ser; n. G2Ser; o. G1Cys; p. G2Cys.
| Species | ABEEMσπ PFF | OPLS/AA | AMBER OL15 |
|---|---|---|---|
| Bond length/nm | 0.0015 | 0.0019 | 0.0019 |
| Angle/(°) | 1.6 | 2.2 | 1.9 |
| Torsion/(°) | 2.3 | 3.0 | 2.8 |
Table 6 RMSDs of geometric structure between force fields and M06-2X/6-311++G** method
| Species | ABEEMσπ PFF | OPLS/AA | AMBER OL15 |
|---|---|---|---|
| Bond length/nm | 0.0015 | 0.0019 | 0.0019 |
| Angle/(°) | 1.6 | 2.2 | 1.9 |
| Torsion/(°) | 2.3 | 3.0 | 2.8 |
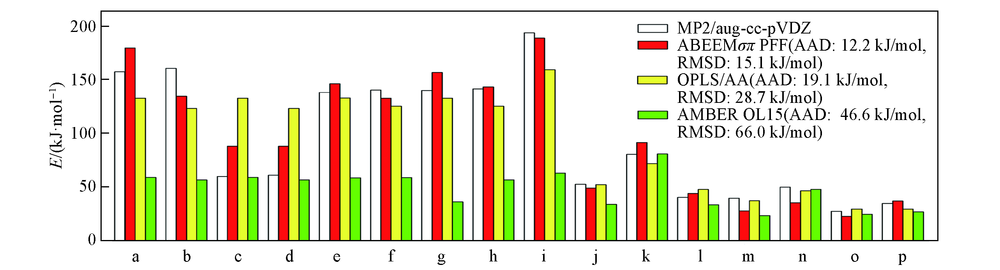
Fig.8 Hydrogen bond energies of QM and various force fieldsa. G3Arg1; b. G3Arg2; c. G4Arg1; d. G4Arg2; e. G5Arg1; f. G5Arg2; g. G6Arg1; h. G6Arg2; i. G3Lys; j. G1Asn1; k. G2Asn1; l. G3Asn2; m. G1Ser; n. G2Ser; o. G1Cys; p. G2Cys.
| Complex | Time/s | Complex | Time/s | ||||||
|---|---|---|---|---|---|---|---|---|---|
| MP2/ aug-cc-pVDZ | ABEEMσπ PFF | OPLS/AA | AMBER OL15 | MP2/ aug-cc-pVDZ | ABEEMσπ PFF | OPLS/AA | AMBER OL15 | ||
| G3Arg1 | 30739.2 | 1.1 | 1.0 | 1.0 | G1Lys | 42499.2 | 1.2 | 1.0 | 1.0 |
| G3Arg2 | 28354.2 | 1.0 | 0.9 | 0.9 | G1Asn1 | 22165.8 | 1.0 | 0.9 | 0.9 |
| G4Arg1 | 33514.8 | 1.1 | 1.0 | 1.0 | G2Asn1 | 21605.4 | 1.0 | 0.9 | 0.9 |
| G4Arg2 | 33964.8 | 1.1 | 1.0 | 1.0 | G3Asn2 | 34574.4 | 1.0 | 0.9 | 0.9 |
| G5Arg1 | 32125.8 | 1.0 | 0.9 | 0.9 | G1Ser | 15585.6 | 1.0 | 0.9 | 0.9 |
| G5Arg2 | 33345.2 | 1.1 | 1.0 | 1.0 | G2Ser | 17013.6 | 1.0 | 0.9 | 0.9 |
| G6Arg1 | 33349.7 | 1.1 | 0.9 | 0.9 | G1Cys | 15991.2 | 1.0 | 0.9 | 0.9 |
| G6Arg2 | 33764.9 | 1.0 | 0.9 | 0.9 | G2Cys | 15226.2 | 1.0 | 0.9 | 0.9 |
Table 7 Computational time of single point energy by various methods
| Complex | Time/s | Complex | Time/s | ||||||
|---|---|---|---|---|---|---|---|---|---|
| MP2/ aug-cc-pVDZ | ABEEMσπ PFF | OPLS/AA | AMBER OL15 | MP2/ aug-cc-pVDZ | ABEEMσπ PFF | OPLS/AA | AMBER OL15 | ||
| G3Arg1 | 30739.2 | 1.1 | 1.0 | 1.0 | G1Lys | 42499.2 | 1.2 | 1.0 | 1.0 |
| G3Arg2 | 28354.2 | 1.0 | 0.9 | 0.9 | G1Asn1 | 22165.8 | 1.0 | 0.9 | 0.9 |
| G4Arg1 | 33514.8 | 1.1 | 1.0 | 1.0 | G2Asn1 | 21605.4 | 1.0 | 0.9 | 0.9 |
| G4Arg2 | 33964.8 | 1.1 | 1.0 | 1.0 | G3Asn2 | 34574.4 | 1.0 | 0.9 | 0.9 |
| G5Arg1 | 32125.8 | 1.0 | 0.9 | 0.9 | G1Ser | 15585.6 | 1.0 | 0.9 | 0.9 |
| G5Arg2 | 33345.2 | 1.1 | 1.0 | 1.0 | G2Ser | 17013.6 | 1.0 | 0.9 | 0.9 |
| G6Arg1 | 33349.7 | 1.1 | 0.9 | 0.9 | G1Cys | 15991.2 | 1.0 | 0.9 | 0.9 |
| G6Arg2 | 33764.9 | 1.0 | 0.9 | 0.9 | G2Cys | 15226.2 | 1.0 | 0.9 | 0.9 |
| [1] | Douki T., Berger M., Raoul S., Ravanat J.L., Cadet J., Determination of 8-Oxo-Purines in DNA by HPLC Using Amperometric Detection., Birkhäuser Press, Basel, 1995, 213-224 |
| [2] | Manas E. S., Getahun Z., Wright W. W., DeGradoW. F., Vanderkooi J. M., J. Am. Chem. Soc.,2000, 122(41), 9883-9890 |
| [3] | Shukla A., Barbiellini B., Buslaps T., Suortti P. Z., Phys. Chem.,2001, 215(10), 1315-1321 |
| [4] | Cadet J., Douki T., Ravanat J. L., Photochem. Photobio.,2015, 91(1), 140-155 |
| [5] | Huo H. J., Zhao D. X., Yang Z. Z., Chem. J. Chinese Universities,2011, 32(12), 2877-2884 |
| (霍红杰, 赵东霞, 杨忠志. 高等学校化学学报, 2011, 32(12), 2877-2884) | |
| [6] | Liu C., Zhang Q. H., Gong L. D., Lu L. N., Yang Z. Z., Chem. J. Chinese Universities,2014, 35(12), 2645-2653 |
| (刘翠, 张千慧, 宫利东, 卢丽男, 杨忠志. 高等学校化学学报, 2014, 35(12), 2645-2653 | |
| [7] | Liu C., Li Y., Han B. Y., Gong L. D., Lu L. N., Yang Z. Z., Zhao D. X., J. Chem. Theory Comput.,2017, 13(5), 2098-2111 |
| [8] | Zhang Q. H., Wang Y., Liu C., Yang Z. Z., Acta Chim. Sinica,2014, 72(8), 956-962 |
| (张千慧, 王阳, 刘翠, 杨忠志. 化学学报, 2014, 72(8), 956-962) | |
| [9] | Liu S., Li S. S., Liu D. J., Wang C. S., Acta Phys. Chim. Sin., 2013, 29(12), 2551-2557 |
| (刘硕, 李书实, 刘冬佳, 王长生. 物理化学学报, 2013, 29(12), 2551-2557) | |
| [10] | Wang C. S., Liu P., Yu N., Acta Phys. Chim. Sin., 2013, 29(6), 1173-1182 |
| (王长生, 刘朋, 于楠. 物理化学学报, 2013, 29(6), 1173-1182) | |
| [11] | González J., Baños I., León I., Contreras-Garcia J., Cocinero E. J., Lesarri A., Fernández J. A., Millán J., J. Chem. Theory Comput., 2016, 12(2), 523-534 |
| [12] | Xie W., Gao J., J. Chem. Theory Comput.,2007, 3(6), 1890-1900 |
| [13] | Wang J., Cieplak P., Li J., Cai Q., Hsieh M., Luo R., Duan Y., J. Phys. Chem. B,2012, 116(24), 7088-7101 |
| [14] | Ivani I., Dans P. D., Noy A., Perez A., Faustino L., Hospital A., Walther J., Andrio P., Goni R., Balaceanu A., Portella G., Battistini F., Gelpi J. L., Gonzalez C., Vendruscolo M., Laughton C. A., Harris S. A., Case D. A., Orozco M., Nat. Methods,2016, 13(1), 55-58 |
| [15] | Kaminski G. A., Stern H. A., Berne B. J., Friesner R. A., Cao Y. X., Murphy R. B., Zhou R., Halgren T. A., J. Comput. Chem., 2002, 23(16), 1515-1531 |
| [16] | Jorgensen W. L., Jensen K. P., Alexandrova A. N., J. Chem. Theory Comput., 2007, 3(6), 1987-1992 |
| [17] | Savelyev A., MacKerell J. A. D., J. Phys. Chem. B,2014, 118(24), 6742-6757 |
| [18] | Lemkul J. A., Huang J., Roux B., Mackerell A. D. Jr., Chem. Rev., 2016, 116(9), 4983-5013 |
| [19] | Zhang C., Bell D., Harger M., Ren P. Y., J. Chem. Theory Comput., 2017, 13(2), 666-678 |
| [20] | Donchev A. G., Ozrin V. D., Subbotin M. V., Tarasov O. V., Tarasov V. I. A., Proc. Natl. Acad. Sci. U. S. A., 2005, 102(22), 7829-7834 |
| [21] | Donchev A. G., Galkin N. G., Illarionov A. A., Khoruzhii O. V., Olevanov M. A., Ozrin V. D., Subbotin M. V., Tarasov V. I., Proc. Natl. Acad. Sci. U. S. A. ,2006, 103(23), 8613-8617 |
| [22] | Zhao D. X., Liu C., Wang F. F., Yu C. Y., Gong L. D., Liu S. B., Yang Z. Z., J. Chem. Theory Comput., 2010, 6(3), 795-804 |
| [23] | Wu Y., Yang Z. Z., J. Phys. Chem. A,2004, 108(37), 7563-7576 |
| [24] | Li X., Yang Z. Z., J. Phys. Chem. A,2005, 109(18), 4102-4111 |
| [25] | Yang Z. Z., Wang J. J., Zhao D. X., J. Comput. Chem., 2014, 35(23), 1690-1706 |
| [26] | Frisch M.J., Trucks G. W., Schlegel H. B., Scuseria G. E., Robb M. A., Cheeseman J. R., Scalmani G., Barone V., Mennucci B., Petersson G. A., Nakatsuji H., Caricato M., Li X., Hratchian H. P., Izmaylov A. F., Bloino J., Zheng G., Sonnenberg J. L., Hada M., Ehara M., Toyota K., Fukuda R., Hasegawa J., Ishida M., Nakajima T., Honda Y., Kitao O., Nakai H., Vreven T., Montgomery J. A., Peralta J. E., Ogliaro F., Bearpark M., Heyd J. J., Brothers E., Kudin K. N., Staroverov V. N., Kobayashi R., Normand J., Raghavachari K., Rendell A., Burant J. C., Iyengar S. S., Tomasi J., Cossi M., Rega N., Millam J. M., Klene M., Knox J. E., Cross J. B., Bakken V., Adamo C., Jaramillo J., Gomperts R., Stratmann R. E., Yazyev O., Austin A. J., Cammi R., Pomelli C., Ochterski J. W., Martin R. L., Morokuma K., Zakrzewski V. G., Voth G. A., Salvador P., Dannenberg J. J., Dapprich S., Daniels A. D., Farkas O., Foresman J. B., Ortiz J. V., Cioslowski J., Fox D. J., Gaussian 09, Revision A.02, Gaussian Inc., Wallingford CT, 2009 |
| [27] | Hohenstein E. G., Chill S. T., Sherrill C. D., J. Chem. Theory Comput., 2008, 4(12), 1996-2000 |
| [1] | CHEN Jialu, HUANG Shuo. Application of Nanopore Sequencing Technology in the Detection of Nucleic Acid Modifications [J]. Chem. J. Chinese Universities, 2022, 43(Album-4): 20220333. |
| [2] | CUI Wei, ZHAO Deyin, BAI Wenxuan, ZHANG Xiaodong, YU Jiang. CO2 Absorption in Composite of Aprotic Solvent and Iron-based Ionic Liquid [J]. Chem. J. Chinese Universities, 2022, 43(8): 20220120. |
| [3] | WEI Chunhong, JIANG Qian, WANG Panpan, JIANG Chengfa, LIU Yuefeng. Atomic Scale Investigation of Pt Atoms/clusters Promoted Co-catalyzed Fischer-Tropsch Synthesis [J]. Chem. J. Chinese Universities, 2022, 43(8): 20220074. |
| [4] | ZHANG Xinxin, XU Di, WANG Yanqiu, HONG Xinlin, LIU Guoliang, YANG Hengquan. Effect of Mn Promoter on CuFe-based Catalysts for CO2 Hydrogenation to Higher Alcohols [J]. Chem. J. Chinese Universities, 2022, 43(7): 20220187. |
| [5] | ZHOU Leilei, CHENG Haiyang, ZHAO Fengyu. Research Progress of CO2 Hydrogenation over Pd-based Heterogeneous Catalysts [J]. Chem. J. Chinese Universities, 2022, 43(7): 20220279. |
| [6] | SONG Youwei, AN Jiangwei, WANG Zheng, WANG Xuhui, QUAN Yanhong, REN Jun, ZHAO Jinxian. Effects of Ag,Zn,Pd-doping on Catalytic Performance of Copper Catalyst for Selective Hydrogenation of Dimethyl Oxalate [J]. Chem. J. Chinese Universities, 2022, 43(6): 20210842. |
| [7] | JI Shuangqi, JIN Zhao, GUAN Wenna, PAN Xiangyu, GUAN Tong. Preparation and Chromatographic Performance of Mixed-mode Silica Stationary Phase Modified by Double Cationic Ionic Liquid and Octadecyl Group [J]. Chem. J. Chinese Universities, 2022, 43(6): 20220008. |
| [8] | MIN Jing, WANG Liyan. 1H NMR Study on the Conformation of Aromatic Amides Limited by Three-center Hydrogen Bonds [J]. Chem. J. Chinese Universities, 2022, 43(6): 20220084. |
| [9] | TANG Gang, SUN Junjie, ZHANG Dongxin, WU Qiang, ZHANG Hexin, SHEN Haifeng, TAO Yi, LIU Xiuyu. Synthesis of Phosphaphenanthrene-modified Cardanol-based Polyol and Its Performance as Flame Retardant in Rigid Polyurethane Foams [J]. Chem. J. Chinese Universities, 2022, 43(4): 20210847. |
| [10] | HU Huimin, CUI Jing, LIU Dandan, SONG Jiaxin, ZHANG Ning, FAN Xiaoqiang, ZHAO Zhen, KONG Lian, XIAO Xia, XIE Zean. Influence of Different Transition Metal Decoration on the Propane Dehydrogenation Performance over Pt/M-DMSN Catalysts [J]. Chem. J. Chinese Universities, 2022, 43(4): 20210815. |
| [11] | TIAN Zhen, DU Na, LI Haiping, SONG Shue, HOU Wanguo. Points of Zero Charge and Surface Acid-base Reaction Equilibrium Constants of Mg-Al-Ti Layered Double Hydroxides [J]. Chem. J. Chinese Universities, 2022, 43(4): 20210833. |
| [12] | XU Dandan, ZOU Xiucheng, LUO Jing, LIU Ren. Synthesis and Characterization of Phenothiazine-based Schiff Bases as Visible Light Photoinitiators [J]. Chem. J. Chinese Universities, 2022, 43(4): 20210857. |
| [13] | ZHANG Yong, XU Jun, BAO Yu, CUI Shuxun. Quantifying the Degree of Weakening Effect of Nonpolar Organic Solvent on the Strength of Intramolecular Hydrogen Bonding by Single-molecule Force Spectroscopy [J]. Chem. J. Chinese Universities, 2022, 43(4): 20210863. |
| [14] | CUI Shaoli, ZHANG Weijia, SHAO Xueguang, CAI Wensheng. Revealing the Effect of Threonine on the Binding Ability of Antifreeze Proteins with Ice Crystals by Free-energy Calculations [J]. Chem. J. Chinese Universities, 2022, 43(3): 20210838. |
| [15] | ZHANG Xiaoyu, XUE Dongping, DU Yu, JIANG Su, WEI Yifan, YAN Wenfu, XIA Huicong, ZHANG Jianan. MOF-derived Carbon-based Electrocatalysts Confinement Catalyst on O2 Reduction and CO2 Reduction Reactions [J]. Chem. J. Chinese Universities, 2022, 43(3): 20210689. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||