

Chem. J. Chinese Universities ›› 2018, Vol. 39 ›› Issue (5): 1026.doi: 10.7503/cjcu20170547
Previous Articles Next Articles
CAO Hongyu1,2,*, JIN Xiaojun1, GUO Wei1, YU Yaxian1, SHI Longfei3, TANG Qian1,2, ZHENG Xuefang1,2,*
Received:2017-08-10
Online:2018-04-18
Published:2018-04-18
Contact:
CAO Hongyu,ZHENG Xuefang
Supported by:CLC Number:
TrendMD:
CAO Hongyu,JIN Xiaojun,GUO Wei,YU Yaxian,SHI Longfei,TANG Qian,ZHENG Xuefang. Investigation on Binding Interactions Between Extracellular Amino-terminal Domain of GLP-1 Receptor Mutations and GLP-1 by Molecular Dynamics Simulations†[J]. Chem. J. Chinese Universities, 2018, 39(5): 1026.
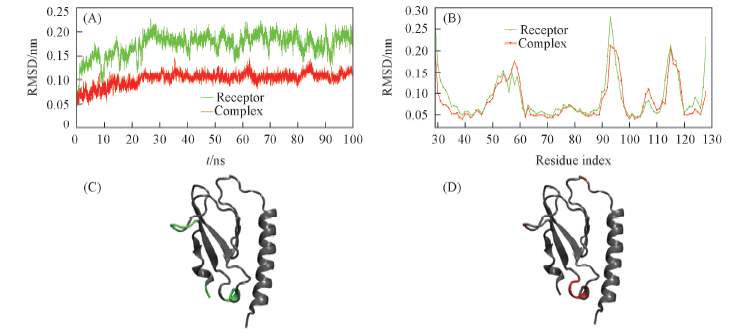
Fig.2 Structual changes of isolated ECD Note: (A) RMSDs of backbone atoms vs. simulation time; (B) RMSF of C-alphas in different residues; (C) unbound ECD structure and residues with RMSF values>0.175 nm, marked in green; (D) ligand-bound ECD structure and residues with RMSF values>0.175 nm, marked in red.
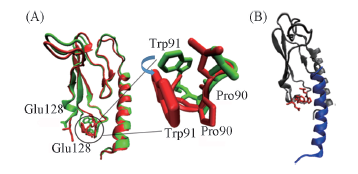
Fig.3 Position changes for residues 89—91 and 128 Note:(A) Superposition of ECDs in the apo form(green) and its ligand-bound form(red) with residues 89—91 and 128 marked in sticks; (B) the schematic plot of ECD(gray) in complex with GLP-1(blue) with residues 89—91 and 128 marked in red.
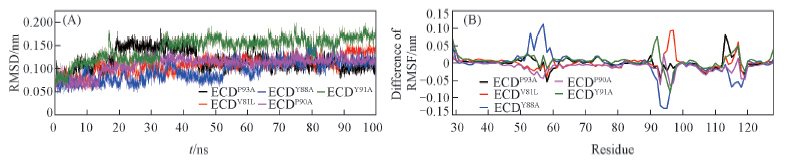
Fig.4 RMSD of ECD mutant backbone and RMSFs of mutant ECD C-Alpha Note:(A) RMSD of ECD mutant backbone versus simulation time; (B) RMSFs of mutant ECD C-alpha.
| Donor | Acceptor | Occupancy(%) | |||||
|---|---|---|---|---|---|---|---|
| WT | ECDP73A | ECDV81L | ECDY88A | ECDP90A | ECDW91A | ||
| Val30-Main | Ser18*-Side | 11.52 | |||||
| Ser31-Side | Glu21*-Side | 64.81 | 42.31 | 65.99 | 34.47 | 78.68 | |
| Leu32-Main | Glu21*-Side | 42.85 | 38.19 | 36.57 | 32.87 | 42.23 | 29.47 |
| Lys26*-Side | Glu128-Side | 100.84 | 99.86 | 92.46 | 76.60 | 102.78 | 89.36 |
| Leu32*-Side | Glu68-Main | 13.28 | 14.74 | 12.54 | 11.26 | 18.22 | 14.14 |
| Arg121-Side | Leu32*-Main | 13.06 | 22.48 | ||||
| Arg121-Side | Val33*-Main | 10.94 | 12.56 | 18.94 | 25.53 | ||
| Lys34*-Side | Glu68-Side | 41.61 | 13.76 | 59.79 | 10.20 | ||
| Lys113-Side | Gly35*-Main | 15.86 | 12.20 | ||||
| Ser116-Side | Gly35*-Main | 11.90 | |||||
| Lys113-Side | Gly35*-Side | 22.44 | 14.26 | 14.72 | 25.57 | ||
| Ser116-Side | Gly35*-Side | 21.32 | 31.23 | ||||
| Ser117-Main | Gly35*-Side | 19.12 | |||||
| Ser117-Side | Gly35*-Side | 17.98 | |||||
| Arg121-Side | Gly35*-Main | 11.78 | |||||
| Arg121-Side | Gly35*-Side | 30.55 | |||||
Table 1 Components of hydrogen bonds between the ECD and GLP-1*
| Donor | Acceptor | Occupancy(%) | |||||
|---|---|---|---|---|---|---|---|
| WT | ECDP73A | ECDV81L | ECDY88A | ECDP90A | ECDW91A | ||
| Val30-Main | Ser18*-Side | 11.52 | |||||
| Ser31-Side | Glu21*-Side | 64.81 | 42.31 | 65.99 | 34.47 | 78.68 | |
| Leu32-Main | Glu21*-Side | 42.85 | 38.19 | 36.57 | 32.87 | 42.23 | 29.47 |
| Lys26*-Side | Glu128-Side | 100.84 | 99.86 | 92.46 | 76.60 | 102.78 | 89.36 |
| Leu32*-Side | Glu68-Main | 13.28 | 14.74 | 12.54 | 11.26 | 18.22 | 14.14 |
| Arg121-Side | Leu32*-Main | 13.06 | 22.48 | ||||
| Arg121-Side | Val33*-Main | 10.94 | 12.56 | 18.94 | 25.53 | ||
| Lys34*-Side | Glu68-Side | 41.61 | 13.76 | 59.79 | 10.20 | ||
| Lys113-Side | Gly35*-Main | 15.86 | 12.20 | ||||
| Ser116-Side | Gly35*-Main | 11.90 | |||||
| Lys113-Side | Gly35*-Side | 22.44 | 14.26 | 14.72 | 25.57 | ||
| Ser116-Side | Gly35*-Side | 21.32 | 31.23 | ||||
| Ser117-Main | Gly35*-Side | 19.12 | |||||
| Ser117-Side | Gly35*-Side | 17.98 | |||||
| Arg121-Side | Gly35*-Main | 11.78 | |||||
| Arg121-Side | Gly35*-Side | 30.55 | |||||
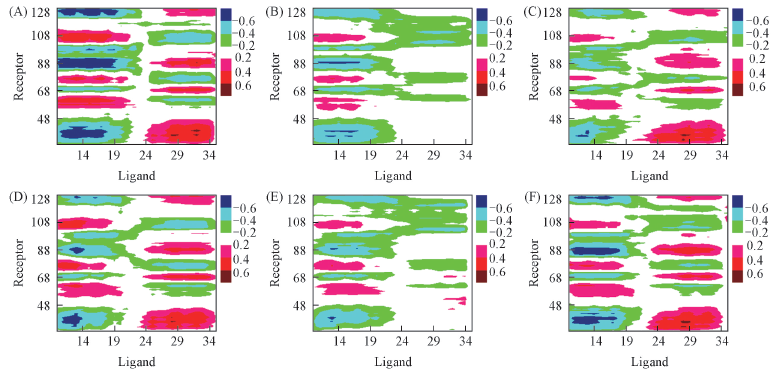
Fig.6 C-Alpha dynamics cross-correlation matrices for residues between the receptor and GLP-1 Note:(A) ECDP73A; (B) ECDV81L; (C) ECDY88A; (D) ECDP90A; (E) ECDW91A; (F) Wild type.
| Residue | Occupancy(%) | Residue | Occupancy(%) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ECDP73A | ECDV81L | ECDY88A | ECDP90A | ECDW91A | WT | ECDP73A | ECDV81L | ECDY88A | ECDP90A | ECDW91A | WT | ||
| S14* | 12 | 1 | 16 | 16 | 3 | 12 | K26* | 100 | 99 | 100 | 99 | 98 | 100 |
| D15* | 9 | 8 | 9 | 10 | 6 | 9 | E27* | 2 | 2 | 2 | 2 | 3 | 2 |
| S17* | 2 | 1 | 7 | 2 | 5 | F28* | 100 | 100 | 100 | 100 | 100 | 100 | |
| S18* | 93 | 95 | 88 | 92 | 92 | 92 | I29* | 100 | 100 | 100 | 100 | 100 | 100 |
| Y19* | 23 | 32 | 29 | 5 | 18 | 18 | A30* | 1 | 1 | 3 | 3 | ||
| E21* | 99 | 100 | 100 | 100 | 100 | 100 | W31* | 78 | 20 | 50 | 87 | 81 | 65 |
| Q23* | 32 | 26 | 44 | 24 | 14 | 26 | L32* | 100 | 100 | 100 | 100 | 100 | 100 |
| A24* | 100 | 100 | 100 | 100 | 100 | 100 | V33* | 100 | 100 | 100 | 100 | 100 | 100 |
| A25* | 100 | 100 | 100 | 100 | 100 | 100 | K34* | 73 | 16 | 99 | 97 | 58 | 41 |
Table 2 Components of interface residues for GLP-1 and their occupancies(%)
| Residue | Occupancy(%) | Residue | Occupancy(%) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ECDP73A | ECDV81L | ECDY88A | ECDP90A | ECDW91A | WT | ECDP73A | ECDV81L | ECDY88A | ECDP90A | ECDW91A | WT | ||
| S14* | 12 | 1 | 16 | 16 | 3 | 12 | K26* | 100 | 99 | 100 | 99 | 98 | 100 |
| D15* | 9 | 8 | 9 | 10 | 6 | 9 | E27* | 2 | 2 | 2 | 2 | 3 | 2 |
| S17* | 2 | 1 | 7 | 2 | 5 | F28* | 100 | 100 | 100 | 100 | 100 | 100 | |
| S18* | 93 | 95 | 88 | 92 | 92 | 92 | I29* | 100 | 100 | 100 | 100 | 100 | 100 |
| Y19* | 23 | 32 | 29 | 5 | 18 | 18 | A30* | 1 | 1 | 3 | 3 | ||
| E21* | 99 | 100 | 100 | 100 | 100 | 100 | W31* | 78 | 20 | 50 | 87 | 81 | 65 |
| Q23* | 32 | 26 | 44 | 24 | 14 | 26 | L32* | 100 | 100 | 100 | 100 | 100 | 100 |
| A24* | 100 | 100 | 100 | 100 | 100 | 100 | V33* | 100 | 100 | 100 | 100 | 100 | 100 |
| A25* | 100 | 100 | 100 | 100 | 100 | 100 | K34* | 73 | 16 | 99 | 97 | 58 | 41 |
| Residue | Occupancy(%) | Residue | Occupancy(%) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ECDP73A | ECDV81L | ECDY88A | ECDP90A | ECDW91A | WT | ECDP73A | ECDV81L | ECDY88A | ECDP90A | ECDW91A | WT | ||
| T29 | 87 | 96 | 95 | 83 | 70 | 96 | L89 | 100 | 100 | 100 | 100 | 100 | 100 |
| V30 | 92 | 90 | 89 | 90 | 100 | 96 | P90 | 94 | 100 | 100 | 17 | 100 | 100 |
| S31 | 95 | 99 | 99 | 97 | 99 | 94 | W91 | 100 | 100 | 100 | 99 | 9 | 100 |
| L32 | 100 | 100 | 100 | 100 | 100 | 100 | A92 | 8 | |||||
| W33 | 60 | 76 | 72 | 62 | 70 | 68 | S93 | 2 | |||||
| E34 | 1 | 4 | 3 | 3 | 4 | 1 | R102 | 2 | 1 | ||||
| T35 | 100 | 100 | 100 | 100 | 100 | 100 | K113 | 43 | 56 | 72 | 12 | 56 | 47 |
| V36 | 100 | 100 | 100 | 100 | 100 | 100 | N115 | 1 | 2 | ||||
| W39 | 100 | 100 | 100 | 100 | 100 | 100 | S116 | 24 | 52 | 4 | 22 | 38 | 46 |
| R40 | 4 | S117 | 8 | 30 | 2 | 13 | 15 | 17 | |||||
| R43 | 2 | 2 | 1 | P118 | 74 | 84 | 99 | 87 | 86 | 97 | |||
| F66 | 1 | P119 | 1 | 1 | 11 | 6 | 25 | ||||||
| D67 | 29 | 15 | 19 | 64 | 27 | 16 | R121 | 100 | 100 | 100 | 100 | 100 | 100 |
| E68 | 100 | 99 | 100 | 100 | 100 | 100 | L123 | 100 | 100 | 100 | 100 | 100 | 100 |
| Y69 | 100 | 100 | 100 | 100 | 100 | 100 | E127 | 36 | 47 | 18 | 21 | 51 | 26 |
| Y88 | 100 | 100 | 49 | 100 | 100 | 100 | E128 | 94 | 88 | 80 | 94 | 86 | 96 |
Table 3 Components of interface residues for GLP-1R ECD and their occupancies(%)
| Residue | Occupancy(%) | Residue | Occupancy(%) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ECDP73A | ECDV81L | ECDY88A | ECDP90A | ECDW91A | WT | ECDP73A | ECDV81L | ECDY88A | ECDP90A | ECDW91A | WT | ||
| T29 | 87 | 96 | 95 | 83 | 70 | 96 | L89 | 100 | 100 | 100 | 100 | 100 | 100 |
| V30 | 92 | 90 | 89 | 90 | 100 | 96 | P90 | 94 | 100 | 100 | 17 | 100 | 100 |
| S31 | 95 | 99 | 99 | 97 | 99 | 94 | W91 | 100 | 100 | 100 | 99 | 9 | 100 |
| L32 | 100 | 100 | 100 | 100 | 100 | 100 | A92 | 8 | |||||
| W33 | 60 | 76 | 72 | 62 | 70 | 68 | S93 | 2 | |||||
| E34 | 1 | 4 | 3 | 3 | 4 | 1 | R102 | 2 | 1 | ||||
| T35 | 100 | 100 | 100 | 100 | 100 | 100 | K113 | 43 | 56 | 72 | 12 | 56 | 47 |
| V36 | 100 | 100 | 100 | 100 | 100 | 100 | N115 | 1 | 2 | ||||
| W39 | 100 | 100 | 100 | 100 | 100 | 100 | S116 | 24 | 52 | 4 | 22 | 38 | 46 |
| R40 | 4 | S117 | 8 | 30 | 2 | 13 | 15 | 17 | |||||
| R43 | 2 | 2 | 1 | P118 | 74 | 84 | 99 | 87 | 86 | 97 | |||
| F66 | 1 | P119 | 1 | 1 | 11 | 6 | 25 | ||||||
| D67 | 29 | 15 | 19 | 64 | 27 | 16 | R121 | 100 | 100 | 100 | 100 | 100 | 100 |
| E68 | 100 | 99 | 100 | 100 | 100 | 100 | L123 | 100 | 100 | 100 | 100 | 100 | 100 |
| Y69 | 100 | 100 | 100 | 100 | 100 | 100 | E127 | 36 | 47 | 18 | 21 | 51 | 26 |
| Y88 | 100 | 100 | 49 | 100 | 100 | 100 | E128 | 94 | 88 | 80 | 94 | 86 | 96 |
| [1] | Lee S.M., Booe J. M., Pioszak A. A., Eur. [J]. Pharmacol., 2015, 763, 196—205 |
| [2] | Miao X.Y., Liu Y., Li C. L., Prog. Biochem. Biophys., 2013, 40(6), 501—509 |
| (苗新宇, 刘瑜, 李春霖. 生物化学与生物物理进展, 2013, 40(6), 501—509) | |
| [3] | Vilsbøll T., Agersø H., Krarup T., Holst J.J., [J]. Clin. Endocrinol. Metabol., 2003, 88(1), 220—224 |
| [4] | Gutniak M.K., Linde B., Holst J. J., Efendi<inline-formula><mml:math xmlns:mml="http://www.w3.org/1998/Math/MathML" id="Mml1-0251-0790-39-5-1026"><mml:mtable frame="none" columnlines="none" rowlines="none"><mml:mtr><mml:mtd><mml:maligngroup/><mml:mrow><mml:mover><mml:mrow><mml:mi mathvariant="normal">c</mml:mi></mml:mrow><mml:mrow><mml:mi mathvariant="normal">'</mml:mi></mml:mrow></mml:mover></mml:mrow></mml:mtd></mml:mtr></mml:mtable></mml:math></inline-formula> S., Diabetes. Care, 1994, 17(9), 1039—1044 |
| [5] | Nie X.H ., Zhang T., Li Y., Zheng X. F., Cao H. Y., Tang Q., Chem. [J]. Chinese Universities, 2010, 31(7), 1337—1341 |
| (聂新华, 张涛, 李元, 郑学仿, 曹洪玉, 唐乾. 高等学校化学学报, 2010, 31(7), 1337—1341) | |
| [6] | De Maturana R.L., Willshaw A., Kuntzsch A., Rudolph R., Donnelly D., [J]. Biol. Chem., 2003, 278(12), 10195—10200 |
| [7] | Runge S., Thøgersen H., Madsen K., Lau J., Rudolph R., [J]. Biol. Chem., 2008, 283(17), 11340—11347 |
| [8] | Underwood C.R., Garibay P., Knudsen L. B., Hastrup S., Peters G. H., Rudolph R., Reedtz R. S., [J]. Biol. Chem., 2010, 285(1), 723—730 |
| [9] | Dods R. L., Donnelly D., Bioscience Reports,2016, 36(1), e00285, 1—13 |
| [10] | Feng X.L., Li Z., Zhao X., Yu H., Liu H. L., Huang X. R., Chem. [J]. Chinese Universities, 2014, 35(2), 338—343 |
| (冯献礼, 李卓, 赵熹, 于辉, 刘慧玲, 黄旭日. 高等学校化学学报, 2014, 35(2), 338—343) | |
| [11] | Cao H. Y., Guo W., Yu Y. X., Wang H. L., Tang Q., Li S. M., Zheng X. F., J. Chem. Theory. Comput., 2017, 16(8), 1750071, 1—15 |
| [12] | Meng X.M., Zhang S. L., Zhang Q. G., Acta Phys. Chim. Sin., 2016, 2, 436—444 |
| (孟现美, 张少龙, 张庆刚. 物理化学学报, 2016, 2, 436—444) | |
| [13] | Huang Y.L., Gao X. F., Chem. [J]. Chinese Universities, 2016, 37(5), 928—931 |
| (黄义玲, 高雪峰. 高等学校化学学报, 2016, 37(5), 928—931) | |
| [14] | Gordon J.C., Myers J. B., Folta T., Shoja V., Heath L. S., Onufriev A., Nucleic Acids Res., 2005, 33(Suppl. 2), W368—W371 |
| [15] | Jo S., Kim T., Iyer V.G., Im W., [J]. Comput. Chem., 2008, 29, 1859—1865 |
| [16] | Jorgensen W.L., Chandrasekhar J., Madura J. D., Impey R. W., Klein M. L., [J]. Chem. Phys., 1983, 79(2), 926—935 |
| [17] | Hess B., Kutzner C., van der Spoel D., Lindahl E., [J]. Chem. Theor. Comp., 2008, 4(3), 435—447 |
| [18] | Hess B., Bekker H., Berendsen H.J., Fraaije J. G. E. M., [J]. Comp. Chem., 1997, 18(12), 1463—1472 |
| [19] | Darden T., York D., Pedersen L., [J]. Chem. Phys., 1993, 98(12), 10089—10092 |
| [20] | Pal K., Melcher K., Xu H.E., Acta Pharmacologica Sinica, 2012, 33(3), 300—311 |
| [21] | Apweiler R., Bairoch A., Wu C.H., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Magrane M., Martin M. J., Natale D. A., O’Donovan C., Redaschi N., Yeh L. S., Nucleic Acids Res., 2004, 32(Suppl. 1), D115—D119 |
| [22] | Simossis V.A., Heringa [J]., Nucleic Acids Res., 2005, 33(Suppl. 2), W289—W294 |
| [23] | Humphrey W., Dalke A., Schulten K., [J]. Mol. Graph., 1996, 14(1), 33—38 |
| [24] | Schymkowitz J., Borg J., Stricher F., Nys R., Rousseau F., Serrano L., Nucleic Acids Res., 2005, 33(Suppl. 2), W382—W388 |
| [25] | Bakan A., Meireles L.M., Bahar I., Bioinformatics, 2011, 27(11), 1575—1577 |
| [26] | Pioszak A.A., Parker N. R., Suino-Powell K., Xu H. E., [J]. Biol. Chem., 2008, 283(47), 32900—32912 |
| [27] | Wilmen A., van Eyll B., Göke B., Göke R., Peptides, 1997, 18(2), 301—305 |
| [1] | GAO Zhiwei, LI Junwei, SHI Sai, FU Qiang, JIA Junru, AN Hailong. Analysis of Gating Characteristics of TRPM8 Channel Based on Molecular Dynamics [J]. Chem. J. Chinese Universities, 2022, 43(6): 20220080. |
| [2] | HU Bo, ZHU Haochen. Dielectric Constant of Confined Water in a Bilayer Graphene Oxide Nanosystem [J]. Chem. J. Chinese Universities, 2022, 43(2): 20210614. |
| [3] | ZHANG Mi, TIAN Yafeng, GAO Keli, HOU Hua, WANG Baoshan. Molecular Dynamics Simulation of the Physicochemical Properties of Trifluoromethanesulfonyl Fluoride Dielectrics [J]. Chem. J. Chinese Universities, 2022, 43(11): 20220424. |
| [4] | LI Congcong, LIU Minghao, HAN Jiarui, ZHU Jingxuan, HAN Weiwei, LI Wannan. Theoretical Study of the Catalytic Activity of VmoLac Non-specific Substrates Based on Molecular Dynamics Simulations [J]. Chem. J. Chinese Universities, 2021, 42(8): 2518. |
| [5] | LEI Xiaotong, JIN Yiqing, MENG Xuanyu. Prediction of the Binding Site of PIP2 in the TREK-1 Channel Based on Molecular Modeling [J]. Chem. J. Chinese Universities, 2021, 42(8): 2550. |
| [6] | ZENG Yonghui, YAN Tianying. Vibrational Density of States Analysis of Proton Hydration Structure [J]. Chem. J. Chinese Universities, 2021, 42(6): 1855. |
| [7] | LIU Aiqing, XU Wensheng, XU Xiaolei, CHEN Jizhong, AN Lijia. Molecular Dynamics Simulation of Polymer/rod Nanocomposite [J]. Chem. J. Chinese Universities, 2021, 42(3): 875. |
| [8] | QI Renrui, LI Minghao, CHANG Hao, FU Xueqi, GAO Bo, HAN Weiwei, HAN Lu, LI Wannan. Theoretical Study on the Unbinding Pathway of Xanthine Oxidase Inhibitors Based on Steered Molecular Dynamics Simulation [J]. Chem. J. Chinese Universities, 2021, 42(3): 758. |
| [9] | QU Siying, XU Qin. Different Roles of Some Key Residues in the S4 Pocket of Coagulation Factor Xa for Rivaroxaban Binding † [J]. Chem. J. Chinese Universities, 2019, 40(9): 1918. |
| [10] | MA Yucong, FAN Baomin, WANG Manman, YANG Biao, HAO Hua, SUN Hui, ZHANG Huijuan. Two-step Preparation of Trazodone and Its Corrosion Inhibition Mechanism for Carbon Steel [J]. Chem. J. Chinese Universities, 2019, 40(8): 1706. |
| [11] | ZHANG Zhang,WANG Dong,WANG Xiaolei,XU Yan. Regulation of Ester Synthesis Activity of Rhizopus chinensis Lipase† [J]. Chem. J. Chinese Universities, 2019, 40(4): 747. |
| [12] | MA Lan,RONG Jingjing,ZHU Youliang,HUANG Yineng,SUN Zhaoyan. Simulation on the Dynamic Process of Formation of Particle Cluster by Generalized Exponential Model† [J]. Chem. J. Chinese Universities, 2019, 40(1): 195. |
| [13] | ZHU Jingxuan,YU Zhengfei,LIU Ye,ZHAN Dongling,HAN Jiarui,TIAN Xiaopian,HAN Weiwei. Exploration of Increasing the Non-specificity Substrates Activity for the Phosphotriesterase-like Lactonase Using Molecular Dynamics Simulations† [J]. Chem. J. Chinese Universities, 2019, 40(1): 138. |
| [14] | WU Hongmei,LI Huiting,LI Yongcheng,WANG Hongqing,WANG Meng. Using Group Contribution Method and Molecular Dynamics to Predict the Glass Transition Temperature of Poly(p-phenylene isophthalamide)† [J]. Chem. J. Chinese Universities, 2019, 40(1): 180. |
| [15] | LIU Yanfang, YANG Hua, ZHANG Hui. Molecular Dynamics Simulation on the Orientation of Alkane Mixture on Graphene† [J]. Chem. J. Chinese Universities, 2018, 39(8): 1729. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||