

Chem. J. Chinese Universities ›› 2019, Vol. 40 ›› Issue (4): 747.doi: 10.7503/cjcu20180437
• Organic Chemistry • Previous Articles Next Articles
ZHANG Zhang1, WANG Dong1,3,*( ), WANG Xiaolei2, XU Yan1,3,*(
), WANG Xiaolei2, XU Yan1,3,*( )
)
Received:2018-06-12
Online:2019-04-03
Published:2018-10-22
Contact:
WANG Dong,XU Yan
E-mail:dwang@jiangnan.edu.cn;yxu@jiangnan.edu.cn
Supported by:CLC Number:
TrendMD:
ZHANG Zhang,WANG Dong,WANG Xiaolei,XU Yan. Regulation of Ester Synthesis Activity of Rhizopus chinensis Lipase†[J]. Chem. J. Chinese Universities, 2019, 40(4): 747.
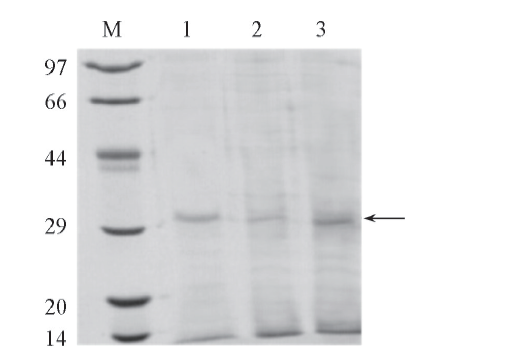
Fig.1 Extraction and isolation of R. chinensis lipase1. The extracted lipase from R. chinensis, eRCL; 2. the supernatant of the extract, eRCL-S; 3. the precipitate of the extract, eRCL-P; M. low molecular protein marker .
| Sample | Concentration/(mg·mL-1) | Hydrolysis activity/(U·mg-1) | Ester synthesis activity /(U·mg-1) |
|---|---|---|---|
| eRCL | 0.75±0.03 | 11.4±0.6 | 10.4±0.6 |
| eRCL-S | 0.35±0.02 | 9.8±0.08 | 8.1±0.04 |
| eRCL-P | 0.98±0.03 | 3.6±0.05 | 1.8±0.11 |
| eRCL-P refolding | 0.22±0.01 | 1.1±0.01 | 0.8±0.01 |
| eRCL-P refolding with eRCL-S | 0.65±0.02 | 8.8±0.06 | 9.1±0.03 |
Table 1 Activity assay of R. chinensis lipase
| Sample | Concentration/(mg·mL-1) | Hydrolysis activity/(U·mg-1) | Ester synthesis activity /(U·mg-1) |
|---|---|---|---|
| eRCL | 0.75±0.03 | 11.4±0.6 | 10.4±0.6 |
| eRCL-S | 0.35±0.02 | 9.8±0.08 | 8.1±0.04 |
| eRCL-P | 0.98±0.03 | 3.6±0.05 | 1.8±0.11 |
| eRCL-P refolding | 0.22±0.01 | 1.1±0.01 | 0.8±0.01 |
| eRCL-P refolding with eRCL-S | 0.65±0.02 | 8.8±0.06 | 9.1±0.03 |

Fig.2 Effect of additives on the activities of R. chinensis lipase r27RCL in refoldingr27RCL, the commercial R. chinensis lipase; MC, membrane component; TX, detergent Triton X-100; with MC, r27RCL refolding with MC; with TX, r27RCL refolding with Triton X-100.
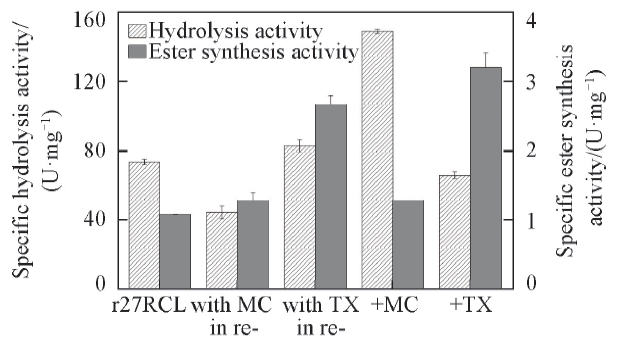
Fig.3 Effect of adding different stages of MC and Triton X-100 on activity of lipase r27RCLMC, membrane component; TX, detergent Triton X-100; with MC in re-, adding MC in renaturation during refolding process; with TX in re-, adding Triton X-100 in renaturation during refolding process; +MC, r27RCL mixed with membrane component directly; +TX, r27RCL mixed with Triton X-100 directly.
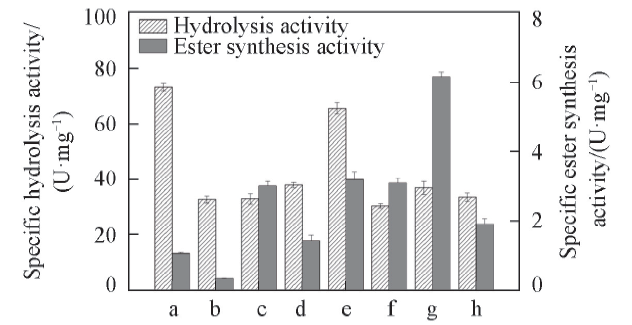
Fig.4 Effects of different detergents on the activity of r27RCLa. r27RCL; b. CTAB; c. Façade-EM; d. C12E8; e. Trion X-100; f. DiC6PC; g. LPC14; h. CHAPS.
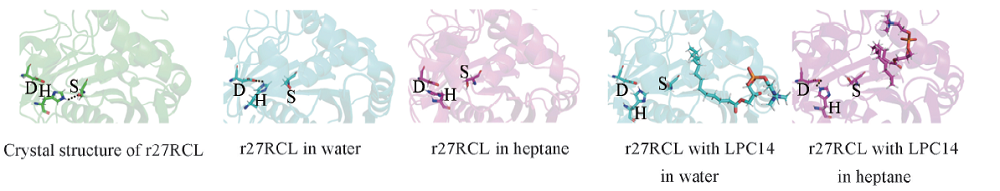
Fig.7 Simulated 3D structure of catalytic triad of r27RCL in different systemsCrystal structure of r27RCL was downloaded from PDB(ID 4L3W) and was showed in green. The simulated 3D structure of r27RCL in water and heptane were showed in cyan and magenta, respectively. The amino acids of catalytic triad of r27RCL and LPC14 were showed as stick.
| [1] | Brzozowski A. M., Derewenda U., Derewenda Z. S., Dodson G. G., Lawson D. M., Turkenburg J. P., Bjorkling F., Huge-Jensen B., Patkar S. A., Thim L., Nature,1991, 351(6326), 491—494 |
| [2] | Jaeger K. E., Eggert T., Curr. Opin.Biotechnol.,2002, 13(4), 390—397 |
| [3] | Cui L. J., Xu G., Meng X., Wu J. P., Yang L. R., Chem. Ind. Eng.Prog.,2014, 33(8), 2150—2154 |
| (崔丽娟, 徐刚, 孟枭, 吴坚平, 杨立荣. 化工进展,2014, 33(8), 2150—2154) | |
| [4] | Jiang L., Yu H., Chem. Res. Chinese Universities,2014, 30(3), 396—399 |
| [5] | Stergiou P. Y., Foukis A., Filippou M., Koukouritaki M., Parapouli M., Theodorou L. G., Hatziloukas E., Afendra A., Pandey A., Papamichael E. M., Biotechnol.Adv.,2013, 31(8), 1846—1859 |
| [6] | Wang Z., Luo W., Fu J., Li Z., Wang Z., Lü P., Chem. Res. Chinese Universities,2017, 33(6), 1—5 |
| [7] | Larios A., García H. S., Oliart R. M., Valerio-Alfaro G., Appl. Microbiol.Biotechnol.,2004, 65(4), 373—376 |
| [8] | Ghanem A., Aboul-Enein H. Y., Tetrahedron:Asymmetry,2004, 15(21), 3331—3351 |
| [9] | Monteiro J. B., Nascimento M. G., Ninow J. L., Biotechnol.Lett.,2003, 25(8), 641—644 |
| [10] | Teng Y., Xu Y., Wang D., J. Mol. Catal. B: Enzym., 2009, 57(1—4), 292—298 |
| [11] | Shah S., Gupta M. N., Process Biochem.,2007, 42(3), 409—414 |
| [12] | Karboune S., Archelas A., Baratti J. C., Process Biochem.,2010, 45(2), 210—216 |
| [13] | Oda M., Kaieda M., Hama S., Yamaji H., Kondo A., Izumoto E., Fukuda H., Biochem. Eng.J.,2005, 23(1), 45—51 |
| [14] | Aarthy M., Saravanan P., Gowthaman M., Rose C., Kamini N., Chem. Eng. Res.Des.,2014, 92(8), 1591—1601 |
| [15] | Xu Y., Wang D., Mu X. Q., Zhao G. A., Zhang K. C., J. Mol. Catal. B: Enzym., 2002, 18(1), 29—37 |
| [16] | Wang D., Xu Y., Teng Y., Bioprocess Biosyst.Eng.,2007, 30(3), 147—155 |
| [17] | He Q., Xu Y., Teng Y., Wang D., Chin. J.Catal.,2008, 29(1), 41—46 |
| [18] | Di Lorenzo M., Hidalgo A., Haas M., Bornscheuer U. T., Appl. Environ.Microbiol.,2005, 71(12), 8974—8977 |
| [19] | Joerger R. D., Haas M. J., Lipids,1993, 28(2), 81—88 |
| [20] | Singh A., Upadhyay V., Upadhyay A. K., Singh S. M., Panda A. K., Microb. Cell.Fact., 2015, 14, 41—51 |
| [21] | Singh S. M., Sharma A., Upadhyay A. K., Singh A., Garg L. C., Panda A. K., Protein Expr.Purif.,2012, 81(1), 75—82 |
| [22] | Satomura A., Kuroda K., Ueda M., PLoS One,2015, 10(5), e0124545 |
| [23] | Akbari N., Khajeh K., Rezaie S., Mirdamadi S., Shavandi M., Ghaemi N., Protein Expr.Purif., 2010, 70(1), 75—80 |
| [24] | Martini V. P., Glogauer A., Müller-Santos M., Iulek J., de Souza E. M., Mitchell D. A., Pedrosa F. O., Krieger N., Microb. Cell.Fact.,2014, 13, 171—185 |
| [25] | Contreras F. X., Ernst A. M., Wieland F., Brugger B., Cold Spring Harb. Perspect. Biol.,2011, 3, 1031—1033 |
| [26] | Laganowsky A., Reading E., Allison T. M., Ulmschneider M. B., Degiacomi M. T., Baldwin A. J., Robinson C. V., Nature,2014, 510(7503), 172—175 |
| [27] | Takada M., Takeuchi H., Shino M., Hamano S., Ohgoh T., Biochem. Biophys. Res.Commun.,1989, 164(2), 653—663 |
| [28] | Yamaguchi H., Miyazaki M., Biomolecules,2014, 4(1), 235—251 |
| [29] | Kordel M., Hofmann B., Schomburg D., Schmid R. D., J.Bacteriol.,1991, 173(15), 4836—4841 |
| [30] | Seddon A. M., Curnow P., Booth P. J., Biochim. Biophys.Acta,2004, 1666(1/2), 105—117 |
| [31] | Otzen D. E., Biophys.J., 2002, 83(4), 2219—2230 |
| [32] | Carrasco-LóPez C., Godoy C., De B. L. R., Fernández-Lorente G., Palomo J. M., Guisán J. M., Fernández-Lafuente R., Martínez-Ripoll M., Hermoso J. A., J. Biol.Chem., 2009, 284(7), 4365—4372 |
| [33] | Tang Q.Y., Wang Y. H.,Mod Food Sci. & Technol., 2017, (3), 179—183 |
| (唐庆芸, 王永华. 现代食品科技, 2017, (3), 179—183) | |
| [34] | Mingarro I., Abad C., Braco L., Proc. Natl. Acad. Sci. USA,1995, 92(8), 3308—3312 |
| [1] | GAO Zhiwei, LI Junwei, SHI Sai, FU Qiang, JIA Junru, AN Hailong. Analysis of Gating Characteristics of TRPM8 Channel Based on Molecular Dynamics [J]. Chem. J. Chinese Universities, 2022, 43(6): 20220080. |
| [2] | HU Bo, ZHU Haochen. Dielectric Constant of Confined Water in a Bilayer Graphene Oxide Nanosystem [J]. Chem. J. Chinese Universities, 2022, 43(2): 20210614. |
| [3] | ZHANG Mi, TIAN Yafeng, GAO Keli, HOU Hua, WANG Baoshan. Molecular Dynamics Simulation of the Physicochemical Properties of Trifluoromethanesulfonyl Fluoride Dielectrics [J]. Chem. J. Chinese Universities, 2022, 43(11): 20220424. |
| [4] | LEI Xiaotong, JIN Yiqing, MENG Xuanyu. Prediction of the Binding Site of PIP2 in the TREK-1 Channel Based on Molecular Modeling [J]. Chem. J. Chinese Universities, 2021, 42(8): 2550. |
| [5] | LI Congcong, LIU Minghao, HAN Jiarui, ZHU Jingxuan, HAN Weiwei, LI Wannan. Theoretical Study of the Catalytic Activity of VmoLac Non-specific Substrates Based on Molecular Dynamics Simulations [J]. Chem. J. Chinese Universities, 2021, 42(8): 2518. |
| [6] | ZENG Yonghui, YAN Tianying. Vibrational Density of States Analysis of Proton Hydration Structure [J]. Chem. J. Chinese Universities, 2021, 42(6): 1855. |
| [7] | QI Renrui, LI Minghao, CHANG Hao, FU Xueqi, GAO Bo, HAN Weiwei, HAN Lu, LI Wannan. Theoretical Study on the Unbinding Pathway of Xanthine Oxidase Inhibitors Based on Steered Molecular Dynamics Simulation [J]. Chem. J. Chinese Universities, 2021, 42(3): 758. |
| [8] | LIU Aiqing, XU Wensheng, XU Xiaolei, CHEN Jizhong, AN Lijia. Molecular Dynamics Simulation of Polymer/rod Nanocomposite [J]. Chem. J. Chinese Universities, 2021, 42(3): 875. |
| [9] | QU Siying, XU Qin. Different Roles of Some Key Residues in the S4 Pocket of Coagulation Factor Xa for Rivaroxaban Binding † [J]. Chem. J. Chinese Universities, 2019, 40(9): 1918. |
| [10] | MA Yucong, FAN Baomin, WANG Manman, YANG Biao, HAO Hua, SUN Hui, ZHANG Huijuan. Two-step Preparation of Trazodone and Its Corrosion Inhibition Mechanism for Carbon Steel [J]. Chem. J. Chinese Universities, 2019, 40(8): 1706. |
| [11] | Xiaoyu FAN,Ke WANG,Shiyong SUN,Biaobiao MA,Rui LÜ. Construction and Catalytic Performances of Fe-aminoclay Nanostructured Lipase † [J]. Chem. J. Chinese Universities, 2019, 40(12): 2512. |
| [12] | ZHU Jingxuan,YU Zhengfei,LIU Ye,ZHAN Dongling,HAN Jiarui,TIAN Xiaopian,HAN Weiwei. Exploration of Increasing the Non-specificity Substrates Activity for the Phosphotriesterase-like Lactonase Using Molecular Dynamics Simulations† [J]. Chem. J. Chinese Universities, 2019, 40(1): 138. |
| [13] | WU Hongmei,LI Huiting,LI Yongcheng,WANG Hongqing,WANG Meng. Using Group Contribution Method and Molecular Dynamics to Predict the Glass Transition Temperature of Poly(p-phenylene isophthalamide)† [J]. Chem. J. Chinese Universities, 2019, 40(1): 180. |
| [14] | MA Lan,RONG Jingjing,ZHU Youliang,HUANG Yineng,SUN Zhaoyan. Simulation on the Dynamic Process of Formation of Particle Cluster by Generalized Exponential Model† [J]. Chem. J. Chinese Universities, 2019, 40(1): 195. |
| [15] | YANG Yihan,WANG Dong,ZHANG Zhang,XU Yan. Activation of Esterification Activity of Rhizopus Chinensis Lipase in Organic Media by Surfactant† [J]. Chem. J. Chinese Universities, 2018, 39(9): 1948. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||