

Chem. J. Chinese Universities ›› 2014, Vol. 35 ›› Issue (2): 338.doi: 10.7503/cjcu20130889
• Physical Chemistry • Previous Articles Next Articles
FENG Xianli1, LI Zhuo1, ZHAO Xi1, YU Hui2, LIU Huiling1, HUANG Xuri1,*( )
)
Received:2013-09-11
Online:2014-02-10
Published:2014-01-02
Contact:
HUANG Xuri
E-mail:huangxr@jlu.edu.cn
Supported by:CLC Number:
TrendMD:
FENG Xianli, LI Zhuo, ZHAO Xi, YU Hui, LIU Huiling, HUANG Xuri. Molecular Dynamics Simulation Study on the Effect of Mutant(Met108→Leu108) on Interactions Between Arabinose-binding Protein and Ligand†[J]. Chem. J. Chinese Universities, 2014, 35(2): 338.
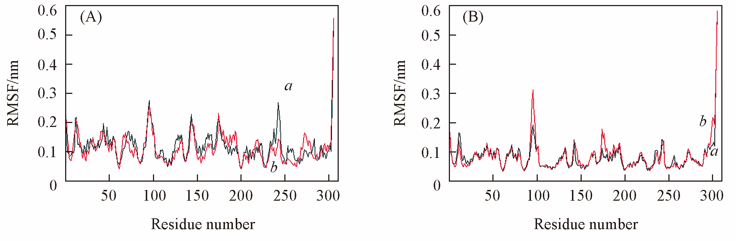
Fig.5 Root mean square fluctuation(RMSF) of the Cα atoms around the average MD structures (A) a. ABP bound ARA; b. ABP bound GAL; (B) a. mutant ABP bound ARA; b. mutant ABP bound GAL.
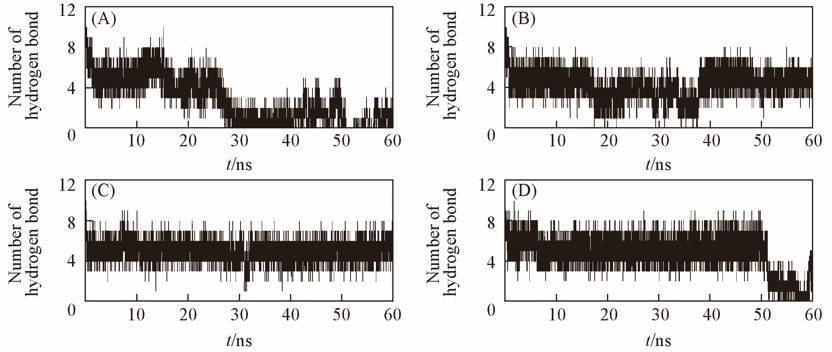
Fig.6 Hydrogen bonding interaction between protein and ligand(A) ABP bound GAL; (B) ABP bound ARA; (C) mutant ABP bound GAL; (D) mutant ABP bound ARA.
| System | Ecoul/(kJ·mol-1) | Evdw/(kJ·mol-1) | ETotal/(kJ·mol-1) |
|---|---|---|---|
| ABP bound GAL | -122.813 | -5.11859 | -127.93 |
| ABP bound ARA | -164.561 | -62.7825 | -227.34 |
| Mutant ABP bound GAL | -173.963 | -78.6312 | -252.59 |
| Mutant ABP bound ARA | -106.336 | -4.034 | -110.37 |
Table 1 Binding free energy components of ABP bound GAL, ABP bound ARA, mutant ABP bound GAL and mutant ABP bound ARA complexes
| System | Ecoul/(kJ·mol-1) | Evdw/(kJ·mol-1) | ETotal/(kJ·mol-1) |
|---|---|---|---|
| ABP bound GAL | -122.813 | -5.11859 | -127.93 |
| ABP bound ARA | -164.561 | -62.7825 | -227.34 |
| Mutant ABP bound GAL | -173.963 | -78.6312 | -252.59 |
| Mutant ABP bound ARA | -106.336 | -4.034 | -110.37 |
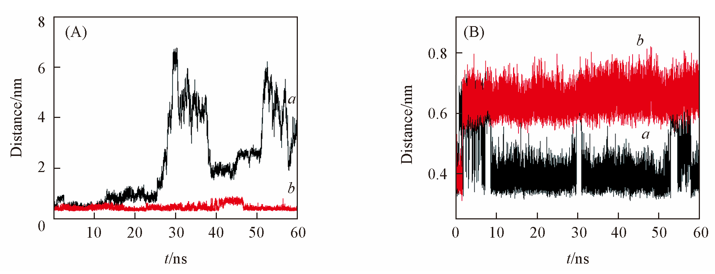
Fig.7 Distance of the pairs of atoms which are specified(A) a. ABP bound ARA, Met108 CE—(GAL C5); b. ABP bound ARA, Met108 CE—(GAL C5); (B) a. mutant ABP bound GAL, Leu108 CD1—(Pro254 CB); b. mutant ABP bOUND ARA, Leu108 CD1—(Pro254 CB).
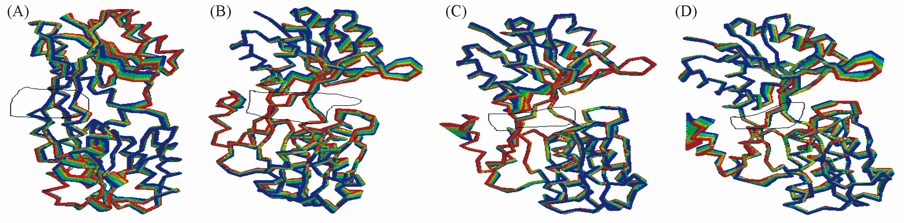
Fig.8 Principal collective motions of the systems of ABP bound ARA(A), ABP bound GAL(B), mutant ABP bound ARA(C) and mutant ABP bound GAL(D) described by the first eigenvector represented as the red structure transforms into the blue structures as it moving in the direction
| [1] | Furlong C.E.; Ed. Neidhardt F. C., Cellular and Molecular Biology, American Society of Microbiology, Washington DC, 1987, 768—796 |
| [2] | Kumar S., Nussinov R., Cell. Mol. Life Sci., 2001, 58, 1216—1233 |
| [3] | Berezovsky N., Shakhnovich E. I., Roc. Natl. Acad. Sci. USA, 2005, 102(36), 12742—12747 |
| [4] | Vieille C., Zeikus G. J., Microbiol. Mol. Biol. Rev., 2001, 65(1), 1—43 |
| [5] | Jaenicke R., Bohm G., Curr. Opin. Struct. Biol., 1998, 8(6), 738—748 |
| [6] | Zhuo K. L., Liu Y. H., Zhang Q. F., Liu H. X., Wang J. J., Chem. J. Chinese Universities, 2008, 29(5), 963—968 |
| (卓克垒, 刘耀辉, 张秋芬, 刘宏勋, 王键吉.高等学校化学学报,2008, 29(5), 963—968) | |
| [7] | Hogg R. W., Englesberg E., J. Bacterial., 1969, 100, 423—432 |
| [8] | Schlief R., J. Mol. Biol., 1969, 46, 185—196 |
| [9] | Miller D. M., Olson J. S., Pflugrath J. W., Quiocho F. A., J. Biol. Chem., 1983, 258, 13665—13672 |
| [10] | Newcomer M. E., Lewis B. A., Quiocho F. A., J. Biol. Chem., 1981, 256, 13218—13222 |
| [11] | Mao B., Pear M. R., McCammon J. A., Quiocho F. A., J. Biol. Chem., 1982, 257, 1131—1133 |
| [12] | Sack J. S., Saper M. A., Quiocho F. A., J. Mol. Biol., 1989, 206, 171—191 |
| [13] | Vermersch P. S., Lemon J., Tesmer J. J., Quiocho F. A., Biochemistry, 1991, 30, 6861—6866 |
| [14] | Quiocho F. A., Vyas N. K., Nature, 1984, 310, 381—386 |
| [15] | Quiocho F. A., Wilson D. K., Vyas N. K., Nature, 1989, 340, 404—407 |
| [16] | Berendsen H. J. C., van der Spoel D., van Drunen R., Computer Physics Communications, 1995, 91, 43—56 |
| [17] | Hornak V., Abel R., Okur A., Strockbine B., Roitberg A., Simmerling C., Structure Function and Structure Function and Bioinformatics, 2006, 65, 712—725 |
| [18] | Hess B., Bekker H., Berendsen H., Fraaije J., J. Comput. Chem., 1997, 18, 1463—1472 |
| [19] | Berendsen H. J. C., Postma J. P. M., van Gunsteren W. F., Dinola A., Haak J. R., J. Chem. Phys., 1984, 81, 3684—3690 |
| [20] | Darden T., York D., Pedersen L., J. Chem. Phys., 1993, 98, 10089—10092 |
| [21] | Essmann U., Perera L., Berkowitz M. L., Darden T., Lee H., Pedersen L. G., J. Chem. Phys., 1995, 103, 8577—8593 |
| [22] | Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., Klein M. L., J. Chem. Phys., 1983, 79, 926—935 |
| [23] | BjoÈrkman A. J., Mowbray S. L., J. Mol. Biol., 1998, 279, 651—664 |
| [1] | GAO Zhiwei, LI Junwei, SHI Sai, FU Qiang, JIA Junru, AN Hailong. Analysis of Gating Characteristics of TRPM8 Channel Based on Molecular Dynamics [J]. Chem. J. Chinese Universities, 2022, 43(6): 20220080. |
| [2] | HU Bo, ZHU Haochen. Dielectric Constant of Confined Water in a Bilayer Graphene Oxide Nanosystem [J]. Chem. J. Chinese Universities, 2022, 43(2): 20210614. |
| [3] | ZHANG Mi, TIAN Yafeng, GAO Keli, HOU Hua, WANG Baoshan. Molecular Dynamics Simulation of the Physicochemical Properties of Trifluoromethanesulfonyl Fluoride Dielectrics [J]. Chem. J. Chinese Universities, 2022, 43(11): 20220424. |
| [4] | LEI Xiaotong, JIN Yiqing, MENG Xuanyu. Prediction of the Binding Site of PIP2 in the TREK-1 Channel Based on Molecular Modeling [J]. Chem. J. Chinese Universities, 2021, 42(8): 2550. |
| [5] | LI Congcong, LIU Minghao, HAN Jiarui, ZHU Jingxuan, HAN Weiwei, LI Wannan. Theoretical Study of the Catalytic Activity of VmoLac Non-specific Substrates Based on Molecular Dynamics Simulations [J]. Chem. J. Chinese Universities, 2021, 42(8): 2518. |
| [6] | ZENG Yonghui, YAN Tianying. Vibrational Density of States Analysis of Proton Hydration Structure [J]. Chem. J. Chinese Universities, 2021, 42(6): 1855. |
| [7] | LIU Aiqing, XU Wensheng, XU Xiaolei, CHEN Jizhong, AN Lijia. Molecular Dynamics Simulation of Polymer/rod Nanocomposite [J]. Chem. J. Chinese Universities, 2021, 42(3): 875. |
| [8] | QI Renrui, LI Minghao, CHANG Hao, FU Xueqi, GAO Bo, HAN Weiwei, HAN Lu, LI Wannan. Theoretical Study on the Unbinding Pathway of Xanthine Oxidase Inhibitors Based on Steered Molecular Dynamics Simulation [J]. Chem. J. Chinese Universities, 2021, 42(3): 758. |
| [9] | QU Siying, XU Qin. Different Roles of Some Key Residues in the S4 Pocket of Coagulation Factor Xa for Rivaroxaban Binding † [J]. Chem. J. Chinese Universities, 2019, 40(9): 1918. |
| [10] | MA Yucong, FAN Baomin, WANG Manman, YANG Biao, HAO Hua, SUN Hui, ZHANG Huijuan. Two-step Preparation of Trazodone and Its Corrosion Inhibition Mechanism for Carbon Steel [J]. Chem. J. Chinese Universities, 2019, 40(8): 1706. |
| [11] | ZHANG Zhang,WANG Dong,WANG Xiaolei,XU Yan. Regulation of Ester Synthesis Activity of Rhizopus chinensis Lipase† [J]. Chem. J. Chinese Universities, 2019, 40(4): 747. |
| [12] | MA Lan,RONG Jingjing,ZHU Youliang,HUANG Yineng,SUN Zhaoyan. Simulation on the Dynamic Process of Formation of Particle Cluster by Generalized Exponential Model† [J]. Chem. J. Chinese Universities, 2019, 40(1): 195. |
| [13] | WU Hongmei,LI Huiting,LI Yongcheng,WANG Hongqing,WANG Meng. Using Group Contribution Method and Molecular Dynamics to Predict the Glass Transition Temperature of Poly(p-phenylene isophthalamide)† [J]. Chem. J. Chinese Universities, 2019, 40(1): 180. |
| [14] | ZHU Jingxuan,YU Zhengfei,LIU Ye,ZHAN Dongling,HAN Jiarui,TIAN Xiaopian,HAN Weiwei. Exploration of Increasing the Non-specificity Substrates Activity for the Phosphotriesterase-like Lactonase Using Molecular Dynamics Simulations† [J]. Chem. J. Chinese Universities, 2019, 40(1): 138. |
| [15] | LIU Yanfang, YANG Hua, ZHANG Hui. Molecular Dynamics Simulation on the Orientation of Alkane Mixture on Graphene† [J]. Chem. J. Chinese Universities, 2018, 39(8): 1729. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||