

高等学校化学学报 ›› 2020, Vol. 41 ›› Issue (11): 2324.doi: 10.7503/cjcu20200405
• 庆祝《高等学校化学学报》复刊40周年专栏 • 上一篇 下一篇
收稿日期:2020-06-30
出版日期:2020-11-10
发布日期:2020-11-06
通讯作者:
黄燕,聂舟
E-mail:yanhuang@hnu.edu.cn;niezhou.hnu@gmail.com
基金资助:
LI Huiyuan, LEI Chunyang, HUANG Yan( ), NIE Zhou(
), NIE Zhou( )
)
Received:2020-06-30
Online:2020-11-10
Published:2020-11-06
Contact:
HUANG Yan,NIE Zhou
E-mail:yanhuang@hnu.edu.cn;niezhou.hnu@gmail.com
Supported by:摘要:
荧光蛋白自发现以来, 因其具有基因编码、 可以自主发出稳健荧光的特点, 在生命科学领域中发挥着重要作用. 随着对荧光蛋白的结构和功能有了更清晰的认识, 在蛋白质工程技术和有机合成迅速发展的基础上, 科研工作者可以对荧光蛋白的结构进行设计改造和模拟, 赋予其新的性质和功能, 扩宽其在生物传感、 生物成像等生命领域的应用. 本文以绿色荧光蛋白的结构改造为主线, 从局部结构改变、 桶状结构重构和表面重构等不同层面阐述了荧光蛋白结构改造的方法以及荧光蛋白模拟物的研究进展, 并介绍了这些荧光蛋白及其模拟物在生物领域的代表性应用.
中图分类号:
TrendMD:
李慧圆, 雷春阳, 黄燕, 聂舟. 荧光蛋白结构改造及其生物传感应用. 高等学校化学学报, 2020, 41(11): 2324.
LI Huiyuan, LEI Chunyang, HUANG Yan, NIE Zhou. Structural Modification of Fluorescent Proteins and Their Applications in Biosensing. Chem. J. Chinese Universities, 2020, 41(11): 2324.
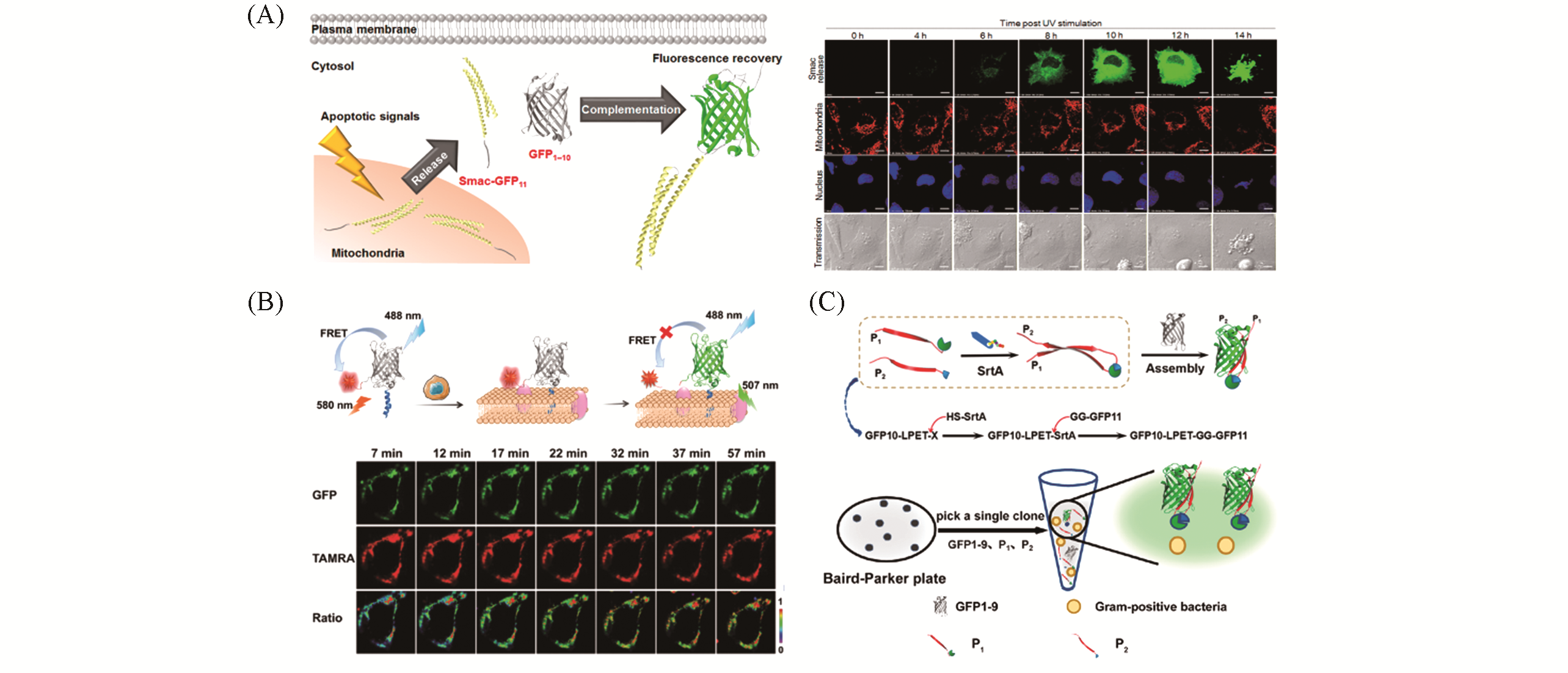
Fig.2 Representative applications of split fluorescent protein in biosensing(A) Schematics for the fluorescence-activatable probe for imaging the release of Smac from mitochondria. Smac-GFP11 is spon- taneously complemented with cytosolic GFP1-10, resulting in the recovery of the fluorescence(left) and time-lapse imaging of Smac release in living cells(right), scale bars: 10 μm[50]; (B) scheme of thesemisynthetic fluorescent protein assembly-based FRET probe(sFPAP) for imaging of furin activity on the living cell membrane(top) and time-lapse imaging of the probe for cell-surface furin assay(bottom)[51]; (C) schematic illustration of Sortase A(SrtA)-catalyzed transpeptidation-mediated assembly of tripartite Split GFP for label-free detection of SrtA activity(top) and schematic illustration of the proposed assay for label-free detection of Gram-positive bacteria(bottom)[56].(A) Copyright 2016, American Chemical Society; (B) Copyright 2019, The Royal Society of Chemistry; (C) Copyright 2018, American Chemical Society.
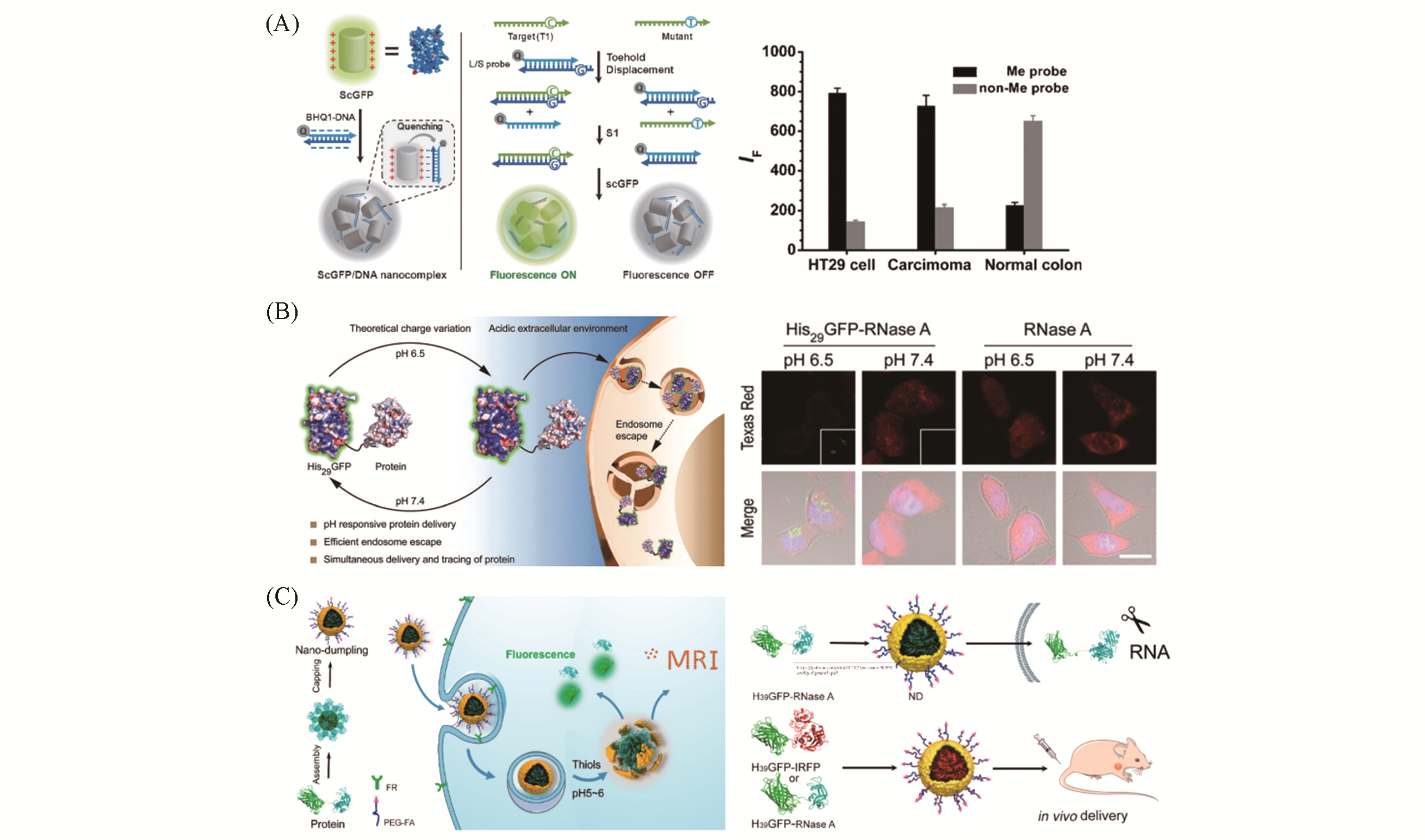
Fig.3 Representative applications of the surface reconstructed fluorescent proteins in biosensing and protein delivery(A) Schematic illustration of the supercharged green fluorescent protein(ScGFP)-based sensing platform for nucleic acid detection and epigenetics analysis based on polyionic nanoscale complex of +36 GFP/DNA and toehold strand displacement(left) and its further application in detection of DNA methylation status in human cancer cell line HT29 and human colon carcinoma tissue sample[61]; (B) pH-responsive delivery and tracing of protein by surface charge designable and tunable GFP(His29GFP)(left) and its further application of delivery of RNase A to HeLa and mRNA content analysis in HeLa by fluorescence in situ hybridization assay(right), scale bar: 20 μm[70]; (C) schematic illustration of cancer cell targeting and fluorescence/MRI bimodal visualized intracellular protein delivery by Protein@InorganicNanodumpling system(NDs)(left) and overview of synthesis and intracellular in vivo delivery of functional protein by NDs(right)[71].(A) Copyright 2014, John Wiley and Sons; (B) Copyright 2018, the Royal Society of Chemistry; (C) Copyright 2020, American Chemical Society.
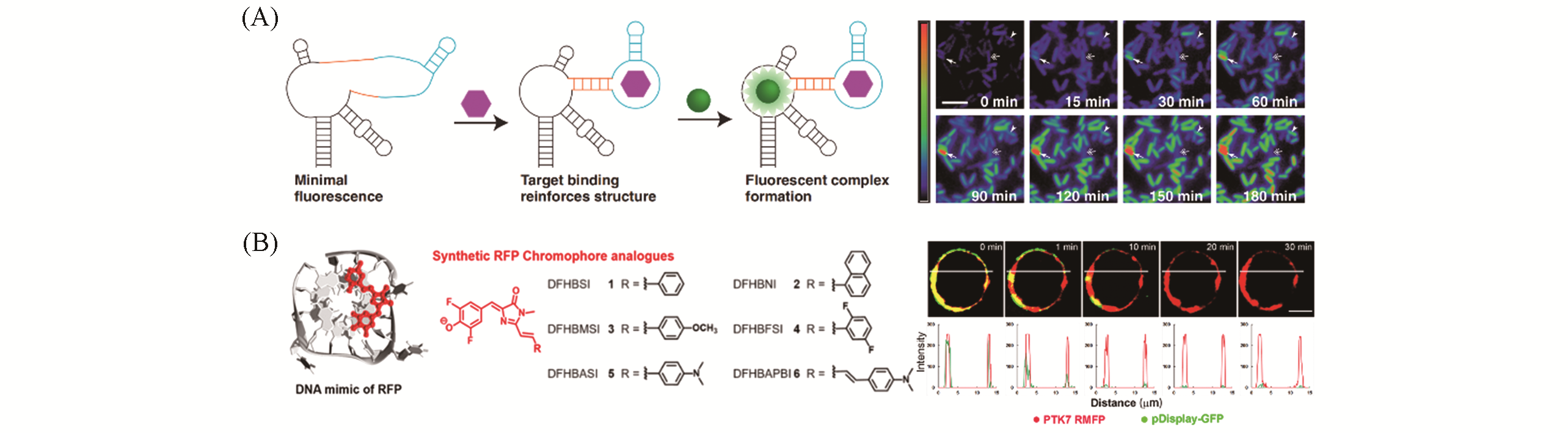
Fig.4 Representative applications of fluorescent protein mimics in biosensing(A) Schematic illustration of the luminous mechanism of an RNA-fluorophore complex “Spinach” sensor(left) and distinct patterns of S-adenosylmethionine(SAM) accumulation after adding methionine to E. coli expressing the SAM sensor RNA(right). Scale bar: 5 μm[87]; (B) structure of a DNA mimic of RFP with the RFP chromophore analogues(left) and its application in bioimaging of the membrane protein PTK(right). Scale bar: 5 μm[101].(A) Copyright 2012, the American Association for the Advancement of Science; (B) Copyright 2017, Oxford University Press.
| 81 | Deng H. P., Zhu X. Y., Mater. Chem. Front., 2017, 1(4), 619—629 |
| 82 | Walker C. L., Lukyanov K. A., Yampolsky I. V., Mishin A. S., Bommarius A. S., Duraj⁃Thatte A. M., Azizi B., Tolbert L. M., Solntsev K. M., Curr. Opin. Chem. Biol., 2015, 27, 64—74 |
| 83 | Babendure J. R., Adams S. R., Tsien R. Y., J. Am. Chem. Soc., 2003, 125(48), 14716—14717 |
| 84 | Murata A., Sato S., Kawazoe Y., Uesugi M., Chem. Commun., 2011, 47(16), 4712—4714 |
| 85 | Sparano B. A., Koide K., J. Am. Chem. Soc., 2005, 127(43), 14954—14955 |
| 86 | Xu W., Lu Y., Anal. Chem., 2010, 82(2), 574—578 |
| 87 | Paige J. S., Nguyen⁃Duc T., Song W., Jaffrey S. R., Science, 2012, 335(6073), 1194 |
| 88 | Kellenberger C. A., Chen C., Whiteley A. T., Portnoy D. A., Hammond M. C., J. Am. Chem. Soc., 2015, 137(20), 6432—6435 |
| 89 | Kellenberger C. A., Wilson S. C., Sales⁃Lee J., Hammond M. C., J. Am. Chem. Soc., 2013, 135(13), 4906—4909 |
| 90 | Nakayama S., Luo Y., Zhou J., Dayie T. K., Sintim H. O., Chem. Commun., 2012, 48(72), 9059—9061 |
| 91 | Song W., Strack R. L., Jaffrey S. R., Nat. Methods, 2013, 10(9), 873—875 |
| 92 | Hofer K., Langejurgen L. V., Jaschke A., J. Am. Chem. Soc., 2013, 135(37), 13692—13694 |
| 93 | Bertucci A., Porchetta A., Ricci F., Anal. Chem., 2018, 90(2), 1049—1053 |
| 94 | You M., Litke J. L., Jaffrey S. R., Proc. Natl. Acad. Sci. USA, 2015, 112(21), E2756—E2765 |
| 95 | Litke J. L., You M., Jaffrey S. R., Methods Enzymol., 2016, 315—333 |
| 96 | Strack R. L., Disney M. D., Jaffrey S. R., Nat. Methods, 2013, 10(12), 1219—1224 |
| 97 | Filonov G. S., Moon J. D., Svensen N., Jaffrey S. R., J. Am. Chem. Soc., 2014, 136(46), 16299—16308 |
| 98 | Song W., Filonov G. S., Kim H., Hirsch M., Li X., Moon J. D., Jaffrey S. R., Nat. Chem. Biol., 2017, 13(11), 1187—1194 |
| 99 | Mudiyanselage A. P. K., Wu R., Leon⁃Duque M. A., Ren K., You M., Methods, 2019, 161, 24—34 |
| 100 | Huang H., Suslov N. B., Li N. S., Shelke S. A., Evans M. E., Koldobskaya Y., Rice P. A., Piccirilli J. A., Nat. Chem. Biol., 2014, 10(8), 686—691 |
| 101 | Feng G., Luo C., Yi H., Yuan L., Lin B., Luo X., Hu X., Wang H., Lei C., Nie Z., Yao S., Nucleic Acids Res., 2017, 45(18), 10380—10392 |
| 1 | Rodriguez E. A., Campbell R. E., Lin J. Y., Lin M. Z., Miyawaki A., Palmer A. E., Shu X., Zhang J., Tsien R. Y., Trends Biochem. Sci., 2017, 42(2), 111—129 |
| 2 | Sanders J. K., Jackson S. E., Chem. Soc. Rev., 2009, 38(10), 2821—2822 |
| 3 | Tsien R. Y., Angew. Chem., 2009, 48(31), 5612—5626 |
| 4 | Davenport D., Nicol J. A. C., Proc. R. Soc. Lond. B Biol. Sci., 1955, 144(916), 399—411 |
| 5 | Shimomura O., Johnson F. H., Saiga Y., J. Cell. Comp. Physiol., 1962, 59(3), 223—239 |
| 6 | Shimomura O., FEBS Lett., 1979, 104(2), 220—222 |
| 7 | Prasher D. C., Eckenrode V. K., Ward W. W., Prendergast F. G., Cormier M. J., Gene, 1992, 111(2), 229—233 |
| 8 | Chalfie M., Tu Y., Euskirchen G., Ward W. W., Prasher D. C. J. S., Science, 1994, 263(5148), 802—805 |
| 9 | Tsien R. Y., Annu. Rev. Biochem., 1998, 67, 509—544 |
| 10 | Matz M. V., Fradkov A. F., Labas Y. A., Savitsky A. P., Zaraisky A. G., Markelov M. L., Lukyanov S. A., Nat. Biotechnol., 1999, 17(10), 969—973 |
| 11 | Campbell R. E., Tour O., Palmer A. E., Steinbach P. A., Baird G. S., Zacharias D. A., Tsien R. Y., Proc. Natl. Acad. Sci. USA, 2002, 99(12), 7877—7882 |
| 12 | Craggs T. D., Chem. Soc. Rev., 2009, 38(10), 2865—2875 |
| 13 | Heim R., Cubitt A. B., Tsien R. Y., Nature, 1995, 373(6516), 663—664 |
| 14 | Heim R., Prasher D. C., Tsien R. Y., Proc. Natl. Acad. Sci. USA, 1994, 91(26), 12501—12504 |
| 15 | Ormö M., Cubitt A. B., Kallio K., Gross L. A., Tsien R. Y., Remington S. J., Science, 1996, 273(5280), 1392—1395 |
| 16 | Rizzuto R., Brini M., De Giorgi F., Rossi R., Heim R., Tsien R. Y., Pozzan T., Curr. Biol., 1996, 6(2), 183—188 |
| 17 | Heim R., Tsien R. Y., Curr. Biol., 1996, 6(2), 178—182 |
| 18 | Llopis J., McCaffery J., Miyawaki A., Llopis J., Heim R., McCaffery J., Adams J., Ikura M., Tsien R., Nature, 1997, 388, 882—887 |
| 19 | Qiao W., Mooney M., Bird A. J., Winge D. R., Eide D. J., Proc. Natl. Acad. Sci. USA, 2006, 103(23), 8674—8679 |
| 20 | Xu X., Gerard A. L., Huang B. C., Anderson D. C., Payan D. G., Luo Y., Nucleic Acids Res., 1998, 26(8), 2034—2035 |
| 21 | Truong K., Sawano A., Mizuno H., Hama H., Tong K. I., Mal T. K., Miyawaki A., Ikura M., Nat. Struct. Biol., 2001, 8(12), 1069—1073 |
| 22 | Zhang J., Allen M. D., Mol. Biosyst., 2007, 3(11), 759—765 |
| 23 | Mattocks J. A., Ho J. V., Cotruvo J. A., J. Am. Chem. Soc., 2019, 141(7), 2857—2861 |
| 24 | Day R. N., Davidson M. W., Chem. Soc. Rev., 2009, 38(10), 2887—2921 |
| 25 | Jackson S. E., Craggs T. D., Huang J. R., Expert Rev. Proteomics, 2006, 3(5), 545—559 |
| 26 | Patterson G. H., Knobel S. M., Sharif W. D., Kain S. R., Piston D. W., Biophys. J., 1997, 73(5), 2782—2790 |
| 27 | Shaner N. C., Steinbach P. A., Tsien R. Y., Nat. Methods, 2005, 2(12), 905—909 |
| 28 | Pedelacq J. D., Cabantous S., Tran T., Terwilliger T. C., Waldo G. S., Nat. Biotechnol., 2006, 24(1), 79—88 |
| 29 | Shaner N. C., Patterson G. H., Davidson M. W., J. Cell Sci., 2007, 120(24), 4247—4260 |
| 30 | Dickson R. M., Cubitt A. B., Tsien R. Y., Moerner W. E., Nature, 1997, 388(6640), 355—358 |
| 31 | Patterson G. H., Lippincott-Schwartz J., Science, 2002, 297(5588), 1873—1877 |
| 32 | Chudakov D. M., Verkhusha V. V., Staroverov D. B., Souslova E. A., Lukyanov S., Lukyanov K. A., Nat. Biotechnol., 2004, 22(11), 1435—1439 |
| 33 | Verkhusha V. V., Sorkin A., Chem. Biol., 2005, 12(3), 279—285 |
| 34 | Subach F. V., Patterson G. H., Manley S., Gillette J. M., Lippincott-Schwartz J., Verkhusha V. V., Nat. Methods, 2009, 6(2), 153—159 |
| 35 | Gurskaya N. G., Verkhusha V. V., Shcheglov A. S., Staroverov D. B., Chepurnykh T. V., Fradkov A. F., Lukyanov S., Lukyanov K. A., Nat. Biotechnol., 2006, 24(4), 461—465 |
| 36 | Johnsson N., Varshavsky A., Proc. Natl. Acad. Sci. USA, 1994, 91(22), 10340—10344 |
| 37 | Rossi F., Charlton C. A., Blau H. M., Proc. Natl. Acad. Sci. USA, 1997, 94(16), 8405—8410 |
| 38 | Pelletier J. N., Campbell-Valois F. X., Michnick S. W., Proc. Natl. Acad. Sci. USA, 1998, 95(21), 12141—12146 |
| 39 | Richards F. M., Proc. Natl. Acad. Sci. USA, 1958, 44(2), 162—166 |
| 40 | Baird G. S., Zacharias D. A., Tsien R. Y., Proc. Natl. Acad. Sci. USA, 1999, 96(20), 11241—11246 |
| 41 | Ghosh I., Hamilton A. D., Regan L., J. Am. Chem. Soc., 2000, 122(23), 5658—5659 |
| 42 | Hu C. D., Chinenov Y., Kerppola T. K., Molecular Cell, 2002, 9(4), 789—798 |
| 43 | Hu C. D., Kerppola T. K., Nat. Biotechnol., 2003, 21(5), 539—545 |
| 44 | Stains C. I., Porter J. R., Ooi A. T., Segal D. J., Ghosh I., J. Am. Chem. Soc., 2005, 127(31), 10782—10783 |
| 45 | Wilson C. G., Magliery T. J., Regan L., Nat. Methods, 2004, 1(3), 255—262 |
| 46 | Kodama Y., Hu C. D., Biotechniques, 2012, 53(5), 285—298 |
| 47 | Pedelacq J. D., Cabantous S., Int. J. Mol. Sci., 2019, 20(14), 3479 |
| 48 | Cabantous S., Terwilliger T. C., Waldo G. S., Nat. Biotechnol., 2005, 23(1), 102—107 |
| 49 | Schmidt S., Adjobo⁃Hermans M. J., Wallbrecher R., Verdurmen W. P., Bovee⁃Geurts P. H., van Oostrum J., Milletti F., Enderle T., Brock R., Angew. Chem., 2015, 54(50), 15105—15108 |
| 50 | Nasu Y., Asaoka Y., Namae M., Nishina H., Yoshimura H., Ozawa T., Anal. Chem., 2016, 88(1), 838—844 |
| 51 | Sun S., Liu Y., Xia J., Wang M., Tang R., Lei C., Huang Y., Nie Z., Yao S., Chem. Commun., 2019, 55(15), 2218—2221 |
| 52 | Yin C., Wang M., Lei C., Wang Z., Li P., Li Y., Li W., Huang Y., Nie Z., Yao S., Anal. Chem., 2015, 87(12), 6311—6318 |
| 53 | Cabantous S., Nguyen H. B., Pedelacq J. D., Koraichi F., Chaudhary A., Ganguly K., Lockard M. A., Favre G., Terwilliger T. C., Waldo G. S., Sci. Rep., 2013, 3, 2854—2863 |
| 54 | Finnigan G. C., Duvalyan A., Liao E. N., Sargsyan A., Thorner J., Mol. Biol. Cell, 2016, 27(17), 2708—2725 |
| 55 | Koraichi F., Gence R., Bouchenot C., Grosjean S., Lajoie⁃Mazenc I., Favre G., Cabantous S., J. Cell Sci., 2018, 131(1), jcs210419 |
| 56 | Zhang J., Wang M., Tang R., Liu Y., Lei C., Huang Y., Nie Z., Yao S., Anal. Chem., 2018, 90(5), 3245—3252 |
| 57 | Zhang Q., Schepis A., Huang H., Yang J., Ma W., Torra J., Zhang S. Q., Yang L., Wu H., Nonell S., Dong Z., Kornberg T. B., Coughlin S. R., Shu X., J. Am. Chem. Soc., 2019, 141(11), 4526—4530 |
| 58 | Gudiksen K. L., Gitlin I., Yang J., Urbach A. R., Moustakas D. T., Whitesides G. M., J. Am. Chem. Soc., 2005, 127(13), 4707—4714 |
| 59 | Lawrence M. S., Phillips K. J., Liu D. R., J. Am. Chem. Soc., 2007, 129(33), 10110—10112 |
| 60 | Thompson D. B., Cronican J. J., Liu D. R., Methods Enzymol, 2012, 503, 293—319 |
| 61 | Lei C., Huang Y., Nie Z., Hu J., Li L., Lu G., Han Y., Yao S., Angew. Chem., 2014, 53(32), 8358—8362 |
| 62 | Wang Z., Li Y., Li L., Li D., Huang Y., Nie Z., Yao S., Chem. Commun., 2015, 51(69), 13373—13376 |
| 63 | Tang S., Nie Z., Li W., Li D., Huang Y., Yao S., Chem. Commun., 2015, 51(76), 14389—14392 |
| 64 | Wang W., Han N., Li R., Han W., Zhang X., Li F., Anal. Chem., 2015, 87(18), 9302—9307 |
| 65 | Wadia J. S., Stan R. V., Dowdy S. F., Nat. Med., 2004, 10(3), 310—315 |
| 66 | Fuchs S. M., Raines R. T., ACS Chem. Biol., 2007, 2(3), 167—170 |
| 67 | McNaughton B. R., Cronican J. J., Thompson D. B., Liu D. R., Proc. Natl. Acad. Sci. USA, 2009, 106(15), 6111—6116 |
| 68 | Cronican J. J., Thompson D. B., Beier K. T., McNaughton B. R., Cepko C. L., Liu D. R., ACS Chem. Biol., 2010, 5(8), 747—752 |
| 69 | Zhao K., Tang Y., Wang Z., Zhang J., Lei C., Wang H., Li H., Huang Y., Nie Z., Yao S., Chem. Commun., 2017, 53(82), 11326—11329 |
| 70 | Hu S., Chen X., Lei C., Tang R., Kang W., Deng H., Huang Y., Nie Z., Yao S., Chem. Commun., 2018, 54(56), 7806—7809 |
| 71 | Zhu X., Tang R., Wang S., Chen X., Hu J., Lei C., Huang Y., Wang H., Nie Z., Yao S., ACS Nano, 2020, 14(2), 2172—2182 |
| 72 | Lei C., Wang Z., Nie Z., Deng H., Hu H., Huang Y., Yao S., Anal. Chem., 2015, 87(3), 1974—1980 |
| 73 | Niwa H., Inouye S., Hirano T., Matsuno T., Kojima S., Kubota M., Ohashi M., Tsuji F. I., Proc. Natl. Acad. Sci. USA, 1996, 93(24), 13617—13622 |
| 74 | Kojima S., Ohkawa H., Hirano T., Maki S., Niwa H., Ohashi M., Inouye S., Tsuji F. I., Tetrahedron Lett., 1998, 39(29), 5239—5242 |
| 75 | Meech S. R.,Chem. Soc. Rev., 2009, 38(10), 2922—2934 |
| 76 | Baldridge A., Samanta S. R., Jayaraj N., Ramamurthy V., Tolbert L. M., J. Am. Chem. Soc., 2010, 132(5), 1498—1499 |
| 77 | Williams D. E., Dolgopolova E. A., Pellechia P. J., Palukoshka A., Wilson T. J., Tan R., Maier J. M., Greytak A. B., Smith M. D., Krause J. A., Shustova N. B., J. Am. Chem. Soc., 2015, 137(6), 2223—2226 |
| 78 | Baldridge A., Feng S., Chang Y. T., Tolbert L. M., ACS Comb. Sci., 2011, 13(3), 214—217 |
| 79 | Povarova N. V., Zaitseva S. O., Baleeva N. S., Smirnov A. Y., Myasnyanko I. N., Zagudaylova M. B., Bozhanova N. G., Gorbachev D. A., Malyshevskaya K. K., Gavrikov A. S., Mishin A. S., Baranov M. S., Chemistry, 2019, 25(41), 9592—9596 |
| 80 | Paige J. S., Wu K. Y., Jaffrey S. R., Science, 2011, 333(6042), 642—646 |
| [1] | 江博文, 陈敬轩, 成永华, 桑微, 寇宗魁. 单原子材料在电化学生物传感中的研究进展[J]. 高等学校化学学报, 2022, 43(9): 20220334. |
| [2] | 王君旸, 刘争, 张茜, 孙春燕, 李红霞. DNA银纳米簇在功能核酸荧光生物传感器中的应用[J]. 高等学校化学学报, 2022, 43(6): 20220010. |
| [3] | 徐梦祎, 黄雪雯, 李小杰, 魏玮, 刘晓亚. “串珠状”复合纳米组装体修饰丝网印刷电极构建的生物传感器[J]. 高等学校化学学报, 2021, 42(6): 1768. |
| [4] | 王博东, 潘美辰, 卓颖. 二氧化硅纳米颗粒表面原位还原银纳米簇电化学发光传感界面的构建与分子识别[J]. 高等学校化学学报, 2021, 42(11): 3519. |
| [5] | 王庆, 何雨秋, 王富安. 多功能脱氧核酶用于生物医学分析的研究进展[J]. 高等学校化学学报, 2021, 42(11): 3334. |
| [6] | 席京, 陈娜, 杨雁冰, 袁荃. 长余辉纳米材料的控制合成及在疾病诊断中的应用[J]. 高等学校化学学报, 2021, 42(11): 3247. |
| [7] | 张嘉懿, 丁臻尧, 王丹丹, 陈礼平, 封心建. 基于多孔金结构的三相界面酶电极的制备及高效电化学酶传感性能[J]. 高等学校化学学报, 2021, 42(10): 3167. |
| [8] | 袁中文, 贺利贞, 陈填烽. 单原子催化剂的生物医学应用[J]. 高等学校化学学报, 2020, 41(12): 2690. |
| [9] | 张怡萌, 张慧欣, 刘洋. 外泌体生物分析及其临床应用研究进展[J]. 高等学校化学学报, 2020, 41(11): 2306. |
| [10] | 郑姗, 刘洋, 陈飘飘, 邢怡晨, 黄朝表. 基于PbS QDs/TiO2 NPs构建新型谷胱甘肽光电化学传感器[J]. 高等学校化学学报, 2019, 40(9): 1866. |
| [11] | 周羽婷, 汤玉娇, 邵爽, 戴诗岩, 程圭芳, 何品刚, 方禹之. 构象转换型传感器对汞、 铅、 锶离子的同时检测[J]. 高等学校化学学报, 2019, 40(8): 1621. |
| [12] | 贾宏亮, 赵建伟, 秦丽溶, 赵敏. 基于镍丝负载氧化镍纳米片的尿酸生物传感器[J]. 高等学校化学学报, 2019, 40(2): 240. |
| [13] | 王春燕,蒋晓青,周泊. 基于Cu-TPA的电化学生物传感器对黄曲霉毒素B1的检测[J]. 高等学校化学学报, 2019, 40(11): 2301. |
| [14] | 贾云静, 史文思, 胡飞柳, 朱华结, 刘莉, 马正月. 漆斑菌中单端孢霉烯化合物及衍生物的细胞毒活性[J]. 高等学校化学学报, 2018, 39(8): 1668. |
| [15] | 黄海平, 岳亚锋, 徐亮, 吕连连, 胡咏梅. 基于Dy2(MoO4)3-AuNPs复合纳米材料的葡萄糖生物传感器[J]. 高等学校化学学报, 2017, 38(4): 554. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||