

Chem. J. Chinese Universities ›› 2018, Vol. 39 ›› Issue (7): 1540.doi: 10.7503/cjcu20170657
• Physical Chemistry • Previous Articles Next Articles
LIU Haichun1, LU Shuai1, ZHANG Yanmin1, ZHOU Weineng1, YIN Lingfeng1, ZHU Lu1, ZHAO Junnan1, LU Tao1,2,*( ), CHEN Yadong1,*(
), CHEN Yadong1,*( )
)
Received:2017-09-29
Online:2018-07-10
Published:2018-06-19
Contact:
LU Tao,CHEN Yadong
E-mail:lutao@cpu.edu.cn;ydchen@cpu.edu.cn
Supported by:CLC Number:
TrendMD:
LIU Haichun, LU Shuai, ZHANG Yanmin, ZHOU Weineng, YIN Lingfeng, ZHU Lu, ZHAO Junnan, LU Tao, CHEN Yadong. Molecular Dynamics Simulation of the Selectivity of Fedratinib Complex with JAK2/JAK3†[J]. Chem. J. Chinese Universities, 2018, 39(7): 1540.
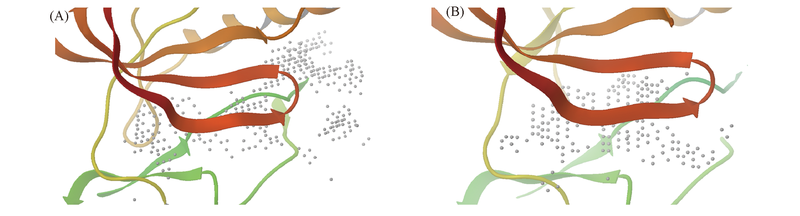
Fig.1 Comparison of “site points” for the ligand binding site in X-ray structures of JAK2(A) and JAK3(B) generated by SiteMap(depicted with white spheres)
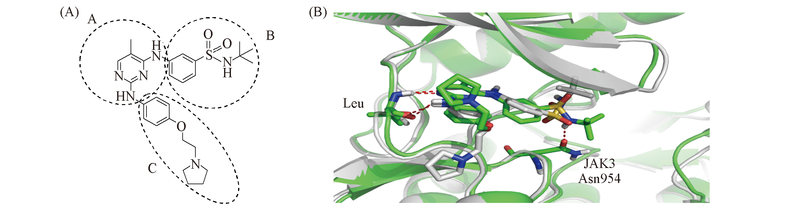
Fig.2 Docking interaction diagram of Fedratinib into the binding site of JAK2(white) and JAK3(green)^ (A) Structure of Fedratinib; (B) binding modes. The hydrogen bonding interactions were depicted with red dotted lines.
| Enzyme | Acceptor | Donor | Distance/nm | Angle/(°) | Occupied(%) |
|---|---|---|---|---|---|
| JAK2 | Fed@O3 | 980@NH2: HH21 | 0.292(±0.014) | 18.58(±9.75) | 99.34 |
| Fed@N2 | 932@N: H | 0.311(±0.014) | 18.14(±9.65) | 98.34 | |
| 932@O | Fed@N3: H2 | 0.302(±0.016) | 30.67(±11.75) | 96.26 | |
| Fed@O2 | 980@NE: HE | 0.311(±0.019) | 35.50(±13.31) | 71.75 | |
| 994@OD1 | Fed@N6: HH27 | 0.305(±0.019) | 21.96(±12.61) | 50.37 | |
| Fed@O2 | 980@NH2: HH21 | 0.329(±0.016) | 47.30(±8.60) | 23.14 | |
| 994@OD2 | Fed@N6: H27 | 0.308(±0.020) | 20.68(±11.45) | 22.30 | |
| Fed@O3 | 980@NE: HE | 0.333(±0.014) | 43.87(±7.05) | 19.98 | |
| JAK3 | Fed@N2 | 905@N: H | 0.314(±0.014) | 17.23(±9.48) | 97.42 |
| 905@O | Fed@N3: H3 | 0.301(±0.016) | 34.29(±12.68) | 92.76 | |
| Fed@O3 | 830@N: H | 0.315(±0.019) | 50.26(±8.20) | 5.42 | |
| Fed@O2 | 855@NZ: HZ1 | 0.328(±0.014) | 43.66(±6.41) | 0.10 |
Table 1 Hydrogen bonds analysis of JAK2/JAK3-Fedratinib from the MD trajectories
| Enzyme | Acceptor | Donor | Distance/nm | Angle/(°) | Occupied(%) |
|---|---|---|---|---|---|
| JAK2 | Fed@O3 | 980@NH2: HH21 | 0.292(±0.014) | 18.58(±9.75) | 99.34 |
| Fed@N2 | 932@N: H | 0.311(±0.014) | 18.14(±9.65) | 98.34 | |
| 932@O | Fed@N3: H2 | 0.302(±0.016) | 30.67(±11.75) | 96.26 | |
| Fed@O2 | 980@NE: HE | 0.311(±0.019) | 35.50(±13.31) | 71.75 | |
| 994@OD1 | Fed@N6: HH27 | 0.305(±0.019) | 21.96(±12.61) | 50.37 | |
| Fed@O2 | 980@NH2: HH21 | 0.329(±0.016) | 47.30(±8.60) | 23.14 | |
| 994@OD2 | Fed@N6: H27 | 0.308(±0.020) | 20.68(±11.45) | 22.30 | |
| Fed@O3 | 980@NE: HE | 0.333(±0.014) | 43.87(±7.05) | 19.98 | |
| JAK3 | Fed@N2 | 905@N: H | 0.314(±0.014) | 17.23(±9.48) | 97.42 |
| 905@O | Fed@N3: H3 | 0.301(±0.016) | 34.29(±12.68) | 92.76 | |
| Fed@O3 | 830@N: H | 0.315(±0.019) | 50.26(±8.20) | 5.42 | |
| Fed@O2 | 855@NZ: HZ1 | 0.328(±0.014) | 43.66(±6.41) | 0.10 |
| Contribution | JAK2 | JAK3 | ||
|---|---|---|---|---|
| Mean/(kJ·mol-1) | Std./(kJ·mol-1) | Mean/(kJ·mol-1) | Std./(kJ·mol-1) | |
| ΔEele/(kJ·mol-1) | -130.79 | 5.14 | -87.99 | 3.23 |
| ΔEvdw/(kJ·mol-1) | -254.09 | 3.26 | -237.19 | 3.35 |
| ΔEintra/(kJ·mol-1) | 0 | 0 | 0 | 0 |
| ΔEgas/(kJ·mol-1) | -384.89 | 5.80 | -325.18 | 4.31 |
| ΔGnp/(kJ·mol-1) | -32.09 | 0.23 | -32.13 | 0.60 |
| ΔGpb/(kJ·mol-1) | 221.67 | 4.35 | 206.35 | 4.21 |
| ΔGpbele/(kJ·mol-1) | 90.88 | 4.57 | 118.41 | 4.22 |
| ΔGpb,tot/(kJ·mol-1) | -195.31 | 5.04 | -150.92 | 4.63 |
| ΔGgb/(kJ·mol-1) | 180.96 | 2.85 | 168.74 | 2.82 |
| ΔGgbele/(kJ·mol-1) | 50.17 | 3.39 | 80.75 | 2.30 |
| ΔGgb,tot/(kJ·mol-1) | -236.02 | 4.23 | -188.53 | 3.26 |
Table 2 Calculated binding free energies of JAK2/JAK3-Fedratinib using MM/GBSA and MM/PBSA method*
| Contribution | JAK2 | JAK3 | ||
|---|---|---|---|---|
| Mean/(kJ·mol-1) | Std./(kJ·mol-1) | Mean/(kJ·mol-1) | Std./(kJ·mol-1) | |
| ΔEele/(kJ·mol-1) | -130.79 | 5.14 | -87.99 | 3.23 |
| ΔEvdw/(kJ·mol-1) | -254.09 | 3.26 | -237.19 | 3.35 |
| ΔEintra/(kJ·mol-1) | 0 | 0 | 0 | 0 |
| ΔEgas/(kJ·mol-1) | -384.89 | 5.80 | -325.18 | 4.31 |
| ΔGnp/(kJ·mol-1) | -32.09 | 0.23 | -32.13 | 0.60 |
| ΔGpb/(kJ·mol-1) | 221.67 | 4.35 | 206.35 | 4.21 |
| ΔGpbele/(kJ·mol-1) | 90.88 | 4.57 | 118.41 | 4.22 |
| ΔGpb,tot/(kJ·mol-1) | -195.31 | 5.04 | -150.92 | 4.63 |
| ΔGgb/(kJ·mol-1) | 180.96 | 2.85 | 168.74 | 2.82 |
| ΔGgbele/(kJ·mol-1) | 50.17 | 3.39 | 80.75 | 2.30 |
| ΔGgb,tot/(kJ·mol-1) | -236.02 | 4.23 | -188.53 | 3.26 |
| Residue | ΔEvdw/(kJ·mol-1) | ΔEele/(kJ·mol-1) | ΔEgas/(kJ·mol-1) | ΔGgb,tot/(kJ·mol-1) | |||||
|---|---|---|---|---|---|---|---|---|---|
| JAK2 | JAK3 | JAK2 | JAK3 | JAK2 | JAK3 | JAK2 | JAK3 | JAK2 | JAK3 |
| Met929 | Met902 | -2.80 | -2.30 | 1.42 | 0.59 | -1.38 | -1.76 | -2.01 | -1.88 |
| Val911 | Val884 | -2.09 | -1.46 | 0.67 | 0.67 | -1.42 | -0.79 | -1.84 | -1.34 |
Table 3 Decomposition of the binding energy on per residue of GateKeeper basis on two systems JAK2-Fedratinib and JAK3-Fedratinib
| Residue | ΔEvdw/(kJ·mol-1) | ΔEele/(kJ·mol-1) | ΔEgas/(kJ·mol-1) | ΔGgb,tot/(kJ·mol-1) | |||||
|---|---|---|---|---|---|---|---|---|---|
| JAK2 | JAK3 | JAK2 | JAK3 | JAK2 | JAK3 | JAK2 | JAK3 | JAK2 | JAK3 |
| Met929 | Met902 | -2.80 | -2.30 | 1.42 | 0.59 | -1.38 | -1.76 | -2.01 | -1.88 |
| Val911 | Val884 | -2.09 | -1.46 | 0.67 | 0.67 | -1.42 | -0.79 | -1.84 | -1.34 |
| Residue | ΔEvdw/(kJ·mol-1) | ΔEele/(kJ·mol-1) | ΔEgas/(kJ·mol-1) | ΔGgb,tot/(kJ·mol-1) | |||||
|---|---|---|---|---|---|---|---|---|---|
| JAK2 | JAK3 | JAK2 | JAK3 | JAK2 | JAK3 | JAK2 | JAK3 | JAK2 | JAK3 |
| Glu930 | Glu903 | -0.71 | -1.38 | -9.29 | -8.79 | -10.00 | -10.17 | -1.67 | -1.42 |
| Leu932 | Leu905 | -5.73 | -5.56 | -12.84 | -11.51 | -18.58 | -17.07 | -11.63 | -10.63 |
Table 4 Decomposition of the binding energy on per residue of hinge region basis on two systems JAK2-Fedratinib and JAK3-Fedratinib
| Residue | ΔEvdw/(kJ·mol-1) | ΔEele/(kJ·mol-1) | ΔEgas/(kJ·mol-1) | ΔGgb,tot/(kJ·mol-1) | |||||
|---|---|---|---|---|---|---|---|---|---|
| JAK2 | JAK3 | JAK2 | JAK3 | JAK2 | JAK3 | JAK2 | JAK3 | JAK2 | JAK3 |
| Glu930 | Glu903 | -0.71 | -1.38 | -9.29 | -8.79 | -10.00 | -10.17 | -1.67 | -1.42 |
| Leu932 | Leu905 | -5.73 | -5.56 | -12.84 | -11.51 | -18.58 | -17.07 | -11.63 | -10.63 |
| Residue | ΔEvdw/(kJ·mol-1) | ΔEele/(kJ·mol-1) | ΔEgas/(kJ·mol-1) | ΔGgb,tot/(kJ·mol-1) | |||||
|---|---|---|---|---|---|---|---|---|---|
| JAK2 | JAK3 | JAK2 | JAK3 | JAK2 | JAK3 | JAK2 | JAK3 | JAK2 | JAK3 |
| Gly856 | Gly829 | -4.23 | -5.10 | -0.79 | -3.05 | -5.02 | -8.20 | -2.85 | -4.73 |
| Lys857 | Lys830 | -3.35 | -5.23 | 3.35 | -10.25 | 0 | -15.44 | -0.21 | -3.31 |
| Gly858 | Gly831 | -1.55 | -3.89 | 0.29 | 4.27 | -1.26 | 0.38 | -0.29 | 0.13 |
| Gly861 | Gly834 | -2.55 | -0.67 | 0.46 | 0.88 | -2.05 | 0.21 | -1.26 | -0.67 |
| Ser862 | Ser835 | -2.59 | -0.75 | -0.54 | -1.21 | -3.14 | -1.97 | -2.59 | -0.96 |
| Val863 | Val836 | -10.38 | -7.32 | 0.04 | 1.46 | -10.33 | -5.82 | -11.88 | -7.91 |
Table 5 Decomposition of the binding energy on per residue of P-loop basis on two systems JAK2-Fedratinib and JAK3-Fedratinib
| Residue | ΔEvdw/(kJ·mol-1) | ΔEele/(kJ·mol-1) | ΔEgas/(kJ·mol-1) | ΔGgb,tot/(kJ·mol-1) | |||||
|---|---|---|---|---|---|---|---|---|---|
| JAK2 | JAK3 | JAK2 | JAK3 | JAK2 | JAK3 | JAK2 | JAK3 | JAK2 | JAK3 |
| Gly856 | Gly829 | -4.23 | -5.10 | -0.79 | -3.05 | -5.02 | -8.20 | -2.85 | -4.73 |
| Lys857 | Lys830 | -3.35 | -5.23 | 3.35 | -10.25 | 0 | -15.44 | -0.21 | -3.31 |
| Gly858 | Gly831 | -1.55 | -3.89 | 0.29 | 4.27 | -1.26 | 0.38 | -0.29 | 0.13 |
| Gly861 | Gly834 | -2.55 | -0.67 | 0.46 | 0.88 | -2.05 | 0.21 | -1.26 | -0.67 |
| Ser862 | Ser835 | -2.59 | -0.75 | -0.54 | -1.21 | -3.14 | -1.97 | -2.59 | -0.96 |
| Val863 | Val836 | -10.38 | -7.32 | 0.04 | 1.46 | -10.33 | -5.82 | -11.88 | -7.91 |
| Residue | ΔEvdw/(kJ·mol-1) | ΔEele/(kJ·mol-1) | ΔEgas/(kJ·mol-1) | ΔGgb,tot/(kJ·mol-1) | |||||
|---|---|---|---|---|---|---|---|---|---|
| JAK2 | JAK3 | JAK2 | JAK3 | JAK2 | JAK3 | JAK2 | JAK3 | JAK2 | JAK3 |
| Lys882 | Lys855 | -3.51 | -1.26 | 12.93 | -6.95 | 9.41 | -8.20 | 3.68 | 1.42 |
| Arg980 | Arg953 | -6.99 | -4.39 | -36.94 | 2.09 | -43.93 | -2.26 | -10.04 | 1.34 |
| Asp994 | Asp967 | -6.82 | -4.39 | -17.20 | 2.64 | -24.02 | -1.76 | -4.98 | -2.38 |
Table 6 Decomposition of the binding energy on some other residue basis on two systems JAK2-Fedratinib and JAK3-Fedratinib
| Residue | ΔEvdw/(kJ·mol-1) | ΔEele/(kJ·mol-1) | ΔEgas/(kJ·mol-1) | ΔGgb,tot/(kJ·mol-1) | |||||
|---|---|---|---|---|---|---|---|---|---|
| JAK2 | JAK3 | JAK2 | JAK3 | JAK2 | JAK3 | JAK2 | JAK3 | JAK2 | JAK3 |
| Lys882 | Lys855 | -3.51 | -1.26 | 12.93 | -6.95 | 9.41 | -8.20 | 3.68 | 1.42 |
| Arg980 | Arg953 | -6.99 | -4.39 | -36.94 | 2.09 | -43.93 | -2.26 | -10.04 | 1.34 |
| Asp994 | Asp967 | -6.82 | -4.39 | -17.20 | 2.64 | -24.02 | -1.76 | -4.98 | -2.38 |
| [1] | Takemoto S., Mulloy J. C., Cereseto A., Migone T. S., Patel B. K. R., Matsuoka M., Yamaguchi K., Takatsuki K., Kamihira S., White J. D., Leonard W. J., Waldmann T., Franchini G., Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 13897—13902 |
| [2] | Ivashkiv L. B., Hu X., Arthritis Rheum., 2003, 48(8), 2092—2096 |
| [3] | Yamaoka K., Saharinen P., Pesu M., Holt V. E. Ⅲ, Silvennoinen O., O’Shea J. J., Genome Biol., 2004, 5(12), 253 |
| [4] | Pesu M., Laurence A., Kishore N., Zwillich S. H., Chan G., O’Shea J. J., Immunol. Rev., 2008, 223, 132—142 |
| [5] | Witthuhn B. A., Quelle F. W., Silvennoinen O., Yi T., Tang B., Miura O., Ihle J. N., Cell, 1993, 74(2), 227—236 |
| [6] | Firmbach-Kraft I., Byers M., Shows T., Dalla-Favera R., Krolewski J. J., Oncogene, 1990, 5(9), 1329—1336 |
| [7] | Harpur A. G., Andres A. C., Ziemiecki A., Aston R. R., Wilks A. F., Oncogene, 1992, 7(7), 1347—1353 |
| [8] | Rane S. G., Reddy E. P., Oncogene, 1994, 9(8), 2415—2423 |
| [9] | Mercier E., Lissalde-Lavigne G.R., Gris J. C., N. Engl. J. Med., 2007, 357, 1984—1985 |
| [10] | Tono C., Xu G., Toki T., Takahashi Y., Sasaki S., Terui K., Ito E., Leukemia, 2005, 19, 1843—1844 |
| [11] | Chen E., Beer P. A., Godfrey A. L., Ortmann C. A., Li J., Costa-Pereira A. P., Ingle C. E., Dermitzakis E. T., Campbell P. J., Green A. R., Cancer Cell, 2010, 18, 524—535 |
| [12] | Bandaranayake R. M., Ungureanu D., Shan Y., Shaw D. E., Silvennoinen O., Hubbard S. R., Nat. Struct. Mol. Biol., 2012, 19(8), 754—759 |
| [13] | Clark J. D., Flanagan M. E., Telliez J. B., J. Med. Chem., 2014, 57(12), 5023—5038 |
| [14] | Menet C. J., Rompaey L. V., Geney R., Prog. Med. Chem., 2013, 52, 153—223 |
| [15] | Zhou T., Georgeon S., Moser R., Moore D. J., Caflisch A., Hantschel O., Leukemia, 2014, 28(2): 404—407 |
| [16] | Kang C. M., Zhao X. H., Yu Y. Q., Lü Y. T., Chem. J. Chinese Universities, 2016, 35(3), 550—554 |
| (康从民, 赵绪浩, 于玉琪, 吕英涛.高等学校化学学报,2016, 35(3), 550—554) | |
| [17] | Wu Y. J., Cui Y. L., Zheng Q. C., Zhang H. X., Chem. J. Chinese Universities, 2014, 35(12), 2605—2611 |
| (吴云剑, 崔颖璐, 郑清川, 张红星.高等学校化学学报,2014, 35(12), 2605—2611) | |
| [18] | Li X. H., Zhao J. W., Teng H., Chem. J. Chinese Universities, 2010, 31(2), 374—378 |
| (李晓晖, 赵俊伟, 滕虎.高等学校化学学报,2010, 31(2), 374—378) | |
| [19] | Zhuang S. L., Wang H. F., Ding K. K., Wang J. Y., Pan L. M., Lu Y. L., Liu Q. J., Zhang C. L., Chemosphere, 2016, 144, 1050—1059 |
| [20] | Muzzioli E., Del Rio A., Rastelli G., Chem. Boil. Drug Des., 2011, 78(2), 252—259 |
| [21] | Berman H. M., Battistuz T., Bhat T. N., Bluhm W. F., Bourne P. E., Burkhardt K., Feng Z., Gilliland G. L., Iype L., Jain S., Fagan P., Marvin J., Padilla D., Ravichandran V., Schneider B., Thanki N., Weissig H., Westbrook J. D., Zardecki C., Acta Crystallogr D, 2002, 58, 899—907 |
| [22] | Maestro, Version 8.5, Schrödinger, LLC. Cambridge, 2008 |
| [23] | Friesner R. A., Banks J. L., Murphy R. B., Halgren T. A., Klicic J. J., Mainz D. T., Repasky M. P., Knoll E. H., Shelley M., Perry J. K., Shaw D. E., Francis P., Shenkin P. S., J. Med. Chem., 2004, 47(7), 1739—1749 |
| [24] | Halgren T. A., Murphy R. B., Friesner R. A., Beard H. S., Frye L. L., Pollard W. T., Banks J. L., J. Med. Chem., 2004, 47(7), 1750—1759 |
| [25] | Case D. A., Cheatham T. E., Darden T., Gohlke H., Luo R., Merz K. M., Onufriev A., Simmerling C., Wang B.,Woods R. J., J. Comput. Chem., 2005, 26(16), 1668—1688 |
| [26] | Frisch M.J., Trucks G. W., Schlegel H. B., Scuseria G. E., Robb M. A., Cheeseman J. R., Scalmani G., Barone V., Mennucci B., Petersson G. A., Nakatsuji H., Caricato M., Li X., Hratchian H. P., Izmaylov A. F., Bloino J., Zheng G., Sonnenberg J. L., Hada M., Ehara M., Toyota K., Fukuda R., Hasegawa J., Ishida M., Nakajima T., Honda Y., Kitao O., Nakai H., Vreven T., Montgomery J. A. Jr., Peralta J. E., Ogliaro F., Bearpark M., Heyd J. J., Brothers E., Kudin K. N., Staroverov V. N., Kobayashi R., Normand J., Raghavachari K., Rendell A., Burant J. C., Iyengar S. S., Tomasi J., Cossi M., Rega N., Millam N. J., Klene M., Knox J. E., Cross J. B., Bakken V., Adamo C., Jaramillo J., Gomperts R., Stratmann R. E., Yazyev O., Austin A. J., Cammi R., Pomelli C., Ochterski J. W., Martin R. L., Morokuma K., Zakrzewski V. G., Voth G. A., Salvador P., Dannenberg J. J., Dapprich S., Daniels A. D., Farkas Ö., Foresman J. B., Ortiz J. V., Cioslowski J., Fox D. J., Gaussian 09, Revision A. 1, Gaussian Inc., Wallingford CT, 2009 |
| [27] | Wang J. M., Wang W., Kollman P. A., Case D. A., J. Mol. Graph. Model., 2006, 25(2), 247—260 |
| [28] | Paschek D., Day R., Garcia A. E., Phys. Chem. Chem. Phys., 2011, 13(44), 19840—19847 |
| [29] | Hou T. J., Wang J. M., Li Y. Y., Wang W., J. Chem. Inf. Model., 2011, 51(1), 69—82 |
| [1] | SONG Yingying, HUANG Lin, LI Qingsen, CHEN Limiao. Preparation of CuO/BiVO4 Photocatalyst and Research on Carbon Dioxide Reduction [J]. Chem. J. Chinese Universities, 2022, 43(6): 20220126. |
| [2] | GAO Zhiwei, LI Junwei, SHI Sai, FU Qiang, JIA Junru, AN Hailong. Analysis of Gating Characteristics of TRPM8 Channel Based on Molecular Dynamics [J]. Chem. J. Chinese Universities, 2022, 43(6): 20220080. |
| [3] | ZENG Xianyang, ZHAO Xi, HUANG Xuri. Mechanism of Inhibition of Glucose and Proton Cotransport Protein GlcPSe by Cytochalasin B [J]. Chem. J. Chinese Universities, 2022, 43(4): 20210822. |
| [4] | CHEN Hanxiang, BIAN Shaoju, HU Bin, LI Wu. Molecular Simulation of the Osmotic Pressures for LiCl-NaCl-KCl-H2O Solution System [J]. Chem. J. Chinese Universities, 2022, 43(3): 20210727. |
| [5] | HU Bo, ZHU Haochen. Dielectric Constant of Confined Water in a Bilayer Graphene Oxide Nanosystem [J]. Chem. J. Chinese Universities, 2022, 43(2): 20210614. |
| [6] | ZHANG Mi, TIAN Yafeng, GAO Keli, HOU Hua, WANG Baoshan. Molecular Dynamics Simulation of the Physicochemical Properties of Trifluoromethanesulfonyl Fluoride Dielectrics [J]. Chem. J. Chinese Universities, 2022, 43(11): 20220424. |
| [7] | ZHANG Lingyu, ZHANG Jilong, QU Zexing. Dynamics Study of Intramolecular Vibrational Energy Redistribution in RDX Molecule [J]. Chem. J. Chinese Universities, 2022, 43(10): 20220393. |
| [8] | LEI Xiaotong, JIN Yiqing, MENG Xuanyu. Prediction of the Binding Site of PIP2 in the TREK-1 Channel Based on Molecular Modeling [J]. Chem. J. Chinese Universities, 2021, 42(8): 2550. |
| [9] | LI Congcong, LIU Minghao, HAN Jiarui, ZHU Jingxuan, HAN Weiwei, LI Wannan. Theoretical Study of the Catalytic Activity of VmoLac Non-specific Substrates Based on Molecular Dynamics Simulations [J]. Chem. J. Chinese Universities, 2021, 42(8): 2518. |
| [10] | SHI Ge, XU Qian, DAI Xiao, ZHANG Jie, SHEN Jun, WAN Xinhua. Effect of Aromatic Substituent on Chiral Recognition of Helical Polyacetylene-based Chiral Stationary Phases for High-Performance Liquid Chromatography [J]. Chem. J. Chinese Universities, 2021, 42(8): 2673. |
| [11] | LIU Shasha, ZHANG Heng, YUAN Shiling, LIU Chengbu. Molecular Dynamics Simulation of Pulsed Electric Field O/W Emulsion Demulsification [J]. Chem. J. Chinese Universities, 2021, 42(7): 2170. |
| [12] | WANG Gaobo, MA Jing. Binding Selectivity Between Diazobenzene and Different Nucleophilic Reagents: Covalent and Noncovalent Interactions [J]. Chem. J. Chinese Universities, 2021, 42(7): 2238. |
| [13] | ZENG Yonghui, YAN Tianying. Vibrational Density of States Analysis of Proton Hydration Structure [J]. Chem. J. Chinese Universities, 2021, 42(6): 1855. |
| [14] | LIU Aiqing, XU Wensheng, XU Xiaolei, CHEN Jizhong, AN Lijia. Molecular Dynamics Simulation of Polymer/rod Nanocomposite [J]. Chem. J. Chinese Universities, 2021, 42(3): 875. |
| [15] | TENG Yunyang, QU Zexing, ZHOU Zhongjun, HUANG Xuri. Theoretical Study on Photoinduced Stepwise Dearomatization of Benzenoid Arenes with Different States [J]. Chem. J. Chinese Universities, 2021, 42(3): 752. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||