

Chem. J. Chinese Universities ›› 2015, Vol. 36 ›› Issue (1): 110.doi: 10.7503/cjcu20140808
• Physical Chemistry • Previous Articles Next Articles
HE Jia1, FENG Xizeng2,3, SHAO Xueguang1,2, CAI Wensheng1,*( )
)
Received:2014-09-05
Revised:2014-12-15
Online:2015-01-10
Published:2014-12-15
Contact:
CAI Wensheng
E-mail:wscai@nankai.edu.cn
Supported by:CLC Number:
TrendMD:
HE Jia, FENG Xizeng, SHAO Xueguang, CAI Wensheng. Adsorption Behavior of Hydrophobin Proteins on Surface of Mica†[J]. Chem. J. Chinese Universities, 2015, 36(1): 110.
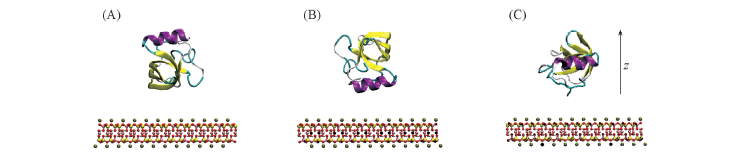
Fig.2 Initial conformations of the molecular systems with three orientations of HFBI towards the surface of mica Three possible orientations are denoted as O1(A), O2(B) and O3(C), indicating the helix of HFBI back to, face to and vertical to the Mica surface. The drawing and coloring style is the same as Fig.1. The water and the ions are omitted for clarity.
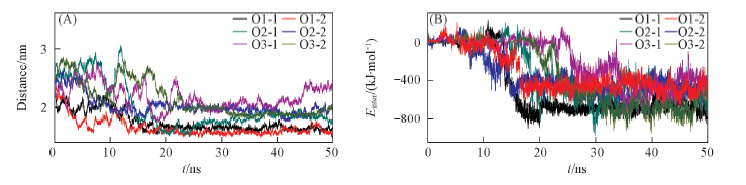
Fig.3 Center of mass distance between HFBI and micas as a function of simulation time(A) and time evolution of interaction energy between HFBI and mica(B)
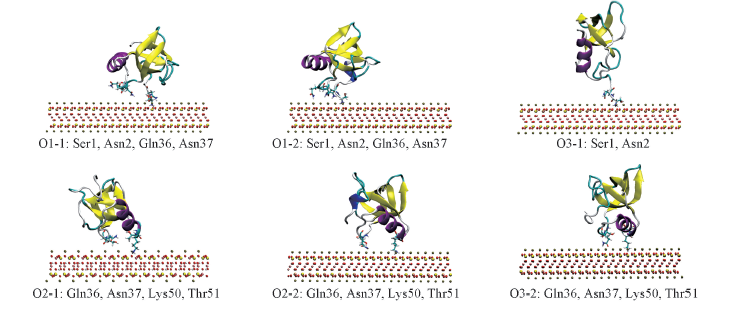
Fig.4 Final conformations of HFBI after the adsorption on Mica The adsorbed patches are clustered into two classes, namely the α-helix(O1-1, O1-2, O3-1) and N-terminus(O2-1, O2-2, O3-2). The adsorbed residues are highlighted in licorice and the rest of drawing and coloring style is the same as Fig.1. The water molecules are omitted for clarity.
| System | RMSD/nm | Rg/nm | Eccentricity | System | RMSD/nm | Rg/nm | Eccentricity |
|---|---|---|---|---|---|---|---|
| O1-1 | 0.19±0.01 | 1.08±0.01 | 0.10±0.01 | O2-2 | 0.19±0.01 | 1.09±0.01 | 0.08±0.03 |
| O1-2 | 0.16±0.01 | 1.08±0.01 | 0.11±0.02 | O3-1 | 0.30±0.02 | 1.12±0.01 | 0.14±0.02 |
| O2-1 | 0.34±0.07 | 1.14±0.01 | 0.10±0.02 | O3-2 | 0.18±0.01 | 1.09±0.01 | 0.07±0.02 |
Table 1 Summary of the simulated systems with averaged HFBI properties by the last 10 ns simulations
| System | RMSD/nm | Rg/nm | Eccentricity | System | RMSD/nm | Rg/nm | Eccentricity |
|---|---|---|---|---|---|---|---|
| O1-1 | 0.19±0.01 | 1.08±0.01 | 0.10±0.01 | O2-2 | 0.19±0.01 | 1.09±0.01 | 0.08±0.03 |
| O1-2 | 0.16±0.01 | 1.08±0.01 | 0.11±0.02 | O3-1 | 0.30±0.02 | 1.12±0.01 | 0.14±0.02 |
| O2-1 | 0.34±0.07 | 1.14±0.01 | 0.10±0.02 | O3-2 | 0.18±0.01 | 1.09±0.01 | 0.07±0.02 |
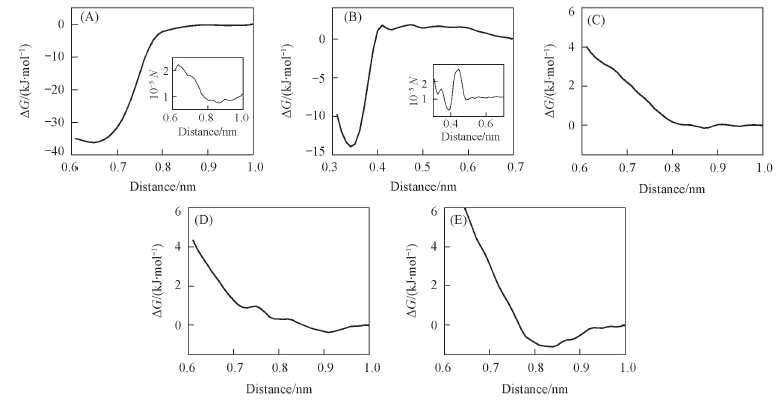
Fig.6 Free-energy profiles for the adsorption of Lys(A), Ser(B), Asn(C), Gln(D) and Thr(E) on the mica surface Inset: Number of samples(N) vs. distance.
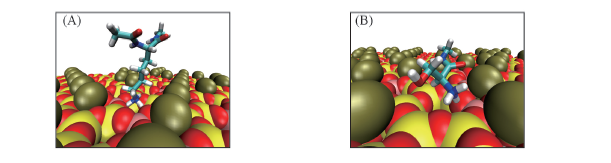
Fig.7 Snapshots of the adsorption structures of Lys(A) and Ser(B) near the global minima of the PMFs The residues are highlighted in licorice. Water and ion molecules are omitted for clarity.
| [1] | Linder M. B., Curr. Opin. Colloid In., 2009, 14(5), 356—363 |
| [2] | Liu Y. Z., Wu M., Feng X. Z., Shao X. G., Cai W. S., J. Phys. Chem. B, 2012, 116(40), 12227—12234 |
| [3] | Liu Y. Z., Cai W. S., Shao X. G., Chem. J. Chinese Universities, 2012, 33(9), 2013—2018 |
| (刘英哲, 蔡文生, 邵学广. 高等学校化学学报, 2012, 33(9), 2013—2018 ) | |
| [4] | Wang L. K., Feng X. Z., Hou S., Chan Q. L., Qin M., Surf. Interface. Anal., 2006, 38, 44—50 |
| [5] | Qin M., Wang L. K., Feng X. Z., Yang Y. L., Wang R., Wang C., Yu L., Shao B., Qiao M. Q., Langmuir , 2007, 23, 4465—4471 |
| [6] | He J., Wu M., Feng X. Z., Shao X. G., Cai W. S., RSC Advances, 2014, 4(26), 13304—13312 |
| [7] | Brancolini G., Kokh D. B., Calzolai L., Wade R. C., Corni S., ACS Nano, 2012, 6, 9863—9878 |
| [8] | Schlegel M. L., Nagy K. L., Fenter P., Cheng L., Sturchio N. C., Jacobsen S. D., Geochim. Cosmochim. Ac., 2006, 70, 3549—3565 |
| [9] | Cheng T., Sun H., J. Phys. Chem. C, 2012, 116, 16436—16446 |
| [10] | Phillips J. C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R. D., Kale L., Schulten K., J. Comput. Chem., 2005, 26(16), 1781—1802 |
| [11] | MacKerell A. D., Bashford D., Bellott M., Dunbrack R. L., Evanseck J. D., Field M. J., Fischer S., Gao J., Guo H., Ha S., Joseph-McCarthy D., Kuchnir L., Kuczera K., Lau F. T. K., Mattos C., Michnick S., Ngo T., Nguyen D. T., Prodhom B., Reiher W. E., Roux B., Schlenkrich M., Smith J. C., Stote R., Straub J., Watanabe M., Wiorkiewicz-Kuczera J., Yin D., Karplus M., J. Phys. Chem. B, 1998, 102(18), 3586—3616 |
| [12] | Mackerell A. D., Feig M., Brooks C. L., J. Comput. Chem., 2004, 25(11), 1400—1415 |
| [13] | Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., Klein M. L., J. Chem. Phys., 1983, 79(2), 926—935 |
| [14] | Heinz H., Koerner H., Anderson K. L., Vaia R. A., Farmer B. L., Chem. Mater., 2005, 17, 5658—5669 |
| [15] | Feller S. E., Zhang Y., Pastor R. W., Brooks B. R., J. Chem. Phys., 1995, 103, 4613—4621 |
| [16] | Tuckerman M., Berne B. J., Martyna G. J., J. Chem. Phys., 1992, 97(3), 1990—2001 |
| [17] | Ryckaert J. P., Ciccotti G., Berendsen H. J. C., J. Comput. Phys., 1977, 23(3), 327—341 |
| [18] | Andersen H. C., J. Comput. Phys., 1983, 52(1), 24—34 |
| [19] | Miyamoto S., Kollman P. A., J. Comput. Chem., 1992, 13(8), 952—962 |
| [20] | Darden T., York D., Pedersen L., J. Chem. Phys., 1993, 98(12), 10089—10092 |
| [21] | Humphrey W., Dalke A., Schulten K., J. Mol. Graphics, 1996, 14(1), 33—38 |
| [22] | Hoefling M., Iori F., Corni S., Gottschalk K. E., Langmuir , 2010, 26(11), 8347—8351 |
| [23] | Darve E., Pohorille A., J. Chem. Phys., 2001, 115, 9169—9183 |
| [24] | Hénin J., Chipot C., J. Chem. Phys., 2004, 121, 2904—2914 |
| [25] | Hénin J., Fiorin G., Chipot C., Klein M. L., J. Chem. Theory Comput., 2010, 6, 35—47 |
| [26] | Fiorin G., Klein M. L., Hénin J., Mol. Phys., 2013, 111, 22—23 |
| [1] | FAN Jianling, TANG Hao, QIN Fengjuan, XU Wenjing, GU Hongfei, PEI Jiajing, CEHN Wenxing. Nitrogen Doped Ultra-thin Carbon Nanosheet Composited Platinum-ruthenium Single Atom Alloy Catalyst for Promoting Electrochemical Hydrogen Evolution Process [J]. Chem. J. Chinese Universities, 2022, 43(9): 20220366. |
| [2] | JIANG Bowen, CHEN Jingxuan, CHENG Yonghua, SANG Wei, KOU Zongkui. Recent Progress of Single-atom Materials in Electrochemical Biosensing [J]. Chem. J. Chinese Universities, 2022, 43(9): 20220334. |
| [3] | WANG Ruyue, WEI Hehe, HUANG Kai, WU Hui. Freezing Synthesis for Single Atom Materials [J]. Chem. J. Chinese Universities, 2022, 43(9): 20220428. |
| [4] | JIANG Shenghan, CAO Changlin, XIAO Liren, YANG Tang, QIAN Qingrong, CHEN Qinghua. Preparation of Composite Semiconductor Micro-sheets with UV Shielding Performance and Its Application in Polypropylene [J]. Chem. J. Chinese Universities, 2022, 43(8): 20220071. |
| [5] | JIN Ruiming, MU Xiaoqing, XU Yan. Bio-chemical Synthesis of Melanin Precursor—— 5,6-Dihydroxyindole(DHI) [J]. Chem. J. Chinese Universities, 2022, 43(8): 20220134. |
| [6] | LI Yulong, XIE Fating, GUAN Yan, LIU Jiali, ZHANG Guiqun, YAO Chao, YANG Tong, YANG Yunhui, HU Rong. A Ratiometric Electrochemical Sensor Based on Silver Ion Interaction with DNA for the Detection of Silver Ion [J]. Chem. J. Chinese Universities, 2022, 43(8): 20220202. |
| [7] | GUO Zhiqiang, YANG Boru, XI Chanjuan. Recent Advances in Reductive Functionalization of Carbon Dioxide with Borohydride Reagents [J]. Chem. J. Chinese Universities, 2022, 43(7): 20220199. |
| [8] | PENG Kuilin, LI Guilin, JIANG Chongyang, ZENG Shaojuan, ZHANG Xiangping. Research Progress for the Role of Electrolytes in the CO2 Electrochemical Reduction [J]. Chem. J. Chinese Universities, 2022, 43(7): 20220238. |
| [9] | WANG Lijun, LI Xin, HONG Song, ZHAN Xinyu, WANG Di, HAO Leiduan, SUN Zhenyu. Efficient Electrocatalytic CO2 Reduction to CO by Tuning CdO-Carbon Black Interface [J]. Chem. J. Chinese Universities, 2022, 43(7): 20220317. |
| [10] | GONG Yanxi, WANG Jianbing, CHAI Buyu, HAN Yuanchun, MA Yunfei, JIA Chaomin. Preparation of Potassium Doped g-C3N4 Thin Film Photoanode and Its Application in Photoelectrocatalytic Oxidation of Diclofenac Sodium in Water [J]. Chem. J. Chinese Universities, 2022, 43(6): 20220005. |
| [11] | GAO Zhiwei, LI Junwei, SHI Sai, FU Qiang, JIA Junru, AN Hailong. Analysis of Gating Characteristics of TRPM8 Channel Based on Molecular Dynamics [J]. Chem. J. Chinese Universities, 2022, 43(6): 20220080. |
| [12] | LIU Qingqing, WANG Pu, WANG Yongshuai, ZHAO Man, DONG Huanli. Synthesis and Topochemical Polymerization Study of Naphthalene/perylene Imides Substituted Diacetylene Derivatives [J]. Chem. J. Chinese Universities, 2022, 43(6): 20220091. |
| [13] | LIU Jiaqi, LI Tianbao. Preparation and Photoelectrochemical Performance of BiVO4/CuBi2O4 Thin Film Photoanodes [J]. Chem. J. Chinese Universities, 2022, 43(4): 20220017. |
| [14] | GUO Jinchang, LIU Fanglin. Planar Pentacoordinate Silicon and Germanium in XBe5H6(X=Si, Ge) Clusters [J]. Chem. J. Chinese Universities, 2022, 43(4): 20210807. |
| [15] | CUI Shaoli, ZHANG Weijia, SHAO Xueguang, CAI Wensheng. Revealing the Effect of Threonine on the Binding Ability of Antifreeze Proteins with Ice Crystals by Free-energy Calculations [J]. Chem. J. Chinese Universities, 2022, 43(3): 20210838. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||