

Chem. J. Chinese Universities ›› 2014, Vol. 35 ›› Issue (4): 776.doi: 10.7503/cjcu20131287
• Physical Chemistry • Previous Articles Next Articles
QI Yanbing, ZHU Jiren, SUN Yaojin, DU Yun, CHU Jianjun, SHI Ting, ZHAO Yilei*( ), WANG Xiaolei*(
), WANG Xiaolei*( )
)
Received:2013-12-27
Online:2014-04-10
Published:2014-02-25
Contact:
ZHAO Yilei,WANG Xiaolei
E-mail:yileizhao@sjtu.edu.cn;thundawner@gmail.com
Supported by:CLC Number:
TrendMD:
QI Yanbing, ZHU Jiren, SUN Yaojin, DU Yun, CHU Jianjun, SHI Ting, ZHAO Yilei, WANG Xiaolei. Theoretical Studies of the Binding-affinity and Reactivity Between Laccase and Phenolic Substrates†[J]. Chem. J. Chinese Universities, 2014, 35(4): 776.
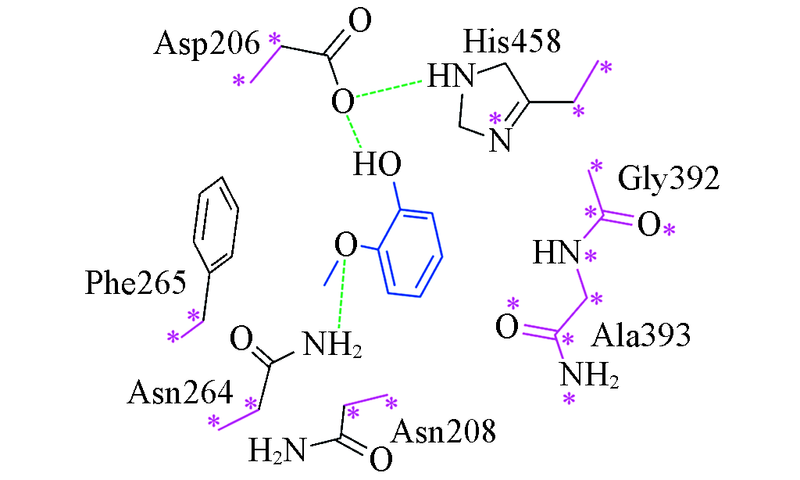
Fig.3 Constructed laccase-substrate model calculated in quantum chemistrySubstrate in blue, constrained heavy atoms in purple and labeled with asterisk, the green dash line represent hydrogen bonding interaction.
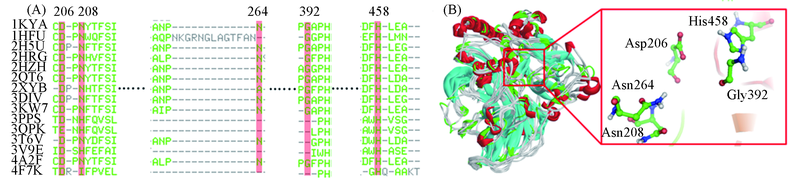
Fig.4 Sequence alignment of the laccases from 15 different organisms(A) and superimposition of the 15 laccase structures and diagram for the common structure of their active pockets(B)
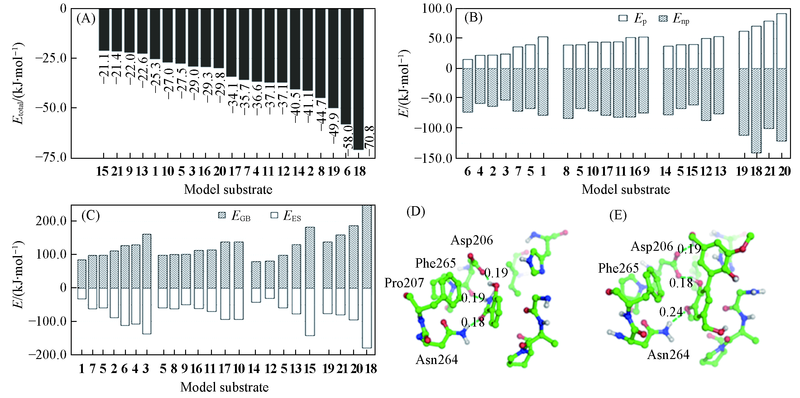
Fig.5 Analysis of binding affinity(A)The total binding energy between laccase and substrates; (B) the polar(Ep) and non-polar(Enp) interaction components;(C) the electrostatic contribution to the solvation free energy calculated by GB(EGB) and electrostatic energy(EES); (D) the complex structure of laccase and substrate 6; (E) the complex structure of laccase and substrate 18. Distance in nm.
| Substrate | ΔEb/(kJ·mol-1) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Leu164 | Asp206 | Asn208 | Phe239 | Asn264 | Phe265 | Pro391 | Gly392 | Ala393 | Pro394 | Hie458 | |
| 1 | -1.34 | 9.45 | -1.34 | -0.42 | -3.51 | -3.55 | -0.29 | -2.80 | 0 | -2.76 | -1.00 |
| 2 | -2.42 | -1.42 | -0.59 | -0.29 | -1.67 | -4.05 | -0.38 | -2.13 | -0.79 | -1.80 | -1.76 |
| 3 | -0.84 | -8.99 | -1.96 | -0.67 | -3.64 | -3.30 | -0.25 | -0.84 | -1.42 | -1.21 | -1.55 |
| 4 | -1.09 | -2.26 | -0.96 | -0.42 | -2.55 | -3.64 | -0.29 | -1.38 | -0.67 | -1.88 | -1.05 |
| 5 | -0.75 | 9.15 | -2.80 | -0.67 | -5.27 | -3.34 | -0.25 | -1.21 | -1.00 | -2.22 | -0.79 |
| 6 | -0.84 | -4.39 | -4.68 | -0.96 | -6.73 | -4.39 | -0.25 | -0.96 | -0.42 | -2.72 | -2.09 |
| 7 | -2.63 | 2.30 | -0.50 | -0.33 | -0.84 | -4.05 | -1.05 | -2.34 | -0.92 | -2.22 | -2.68 |
| 8 | -1.46 | 6.44 | -2.01 | -0.50 | -5.73 | -4.56 | -0.25 | -3.39 | -0.67 | -3.97 | -0.75 |
| 9 | -2.13 | 9.78 | -0.96 | -0.50 | -5.35 | -3.01 | -0.38 | -3.55 | -0.38 | -2.51 | -0.54 |
| 10 | -1.71 | 8.40 | -0.84 | -0.46 | -6.10 | -3.51 | -0.33 | -3.55 | -2.97 | -2.93 | -0.71 |
| 11 | -1.34 | 6.77 | -2.05 | -0.46 | -6.10 | -4.35 | -0.25 | -2.88 | -1.25 | -4.22 | -1.09 |
| 12 | -2.63 | 7.15 | -2.05 | -0.54 | -3.89 | -3.68 | -0.46 | -4.39 | -0.29 | -2.88 | -0.88 |
| 13 | -2.26 | 9.24 | -0.92 | -0.42 | -4.47 | -3.14 | -0.50 | -4.47 | 0.04 | -1.71 | -0.71 |
| 14 | -2.34 | 3.76 | -1.09 | -0.46 | -1.25 | -4.60 | -0.42 | -2.63 | -0.42 | -2.55 | -1.30 |
| 15 | -2.51 | -6.94 | -0.75 | -0.42 | -1.59 | -3.55 | -0.46 | -2.88 | -0.59 | -1.80 | -1.00 |
| 16 | -1.30 | 8.74 | -1.76 | -0.46 | -6.23 | -4.01 | -0.33 | -2.84 | -0.04 | -3.05 | -0.88 |
| 17 | -1.50 | 8.28 | -1.00 | -0.46 | -4.72 | -4.85 | -0.25 | -2.30 | -1.30 | -3.55 | -0.88 |
| 18 | -2.30 | 10.24 | -0.88 | -0.46 | -2.93 | -6.60 | -5.77 | -5.64 | -3.68 | -3.59 | -5.98 |
| 19 | -8.53 | 8.40 | -0.13 | -0.21 | 0.04 | -3.05 | -4.81 | -6.60 | 1.34 | -1.67 | -4.39 |
| 20 | -4.97 | 8.07 | -0.75 | -0.38 | -4.18 | -5.31 | -0.92 | -4.93 | -1.38 | -5.94 | -3.72 |
| 21 | -2.68 | 9.91 | -1.38 | -0.42 | -5.31 | -7.77 | -1.21 | -3.68 | -3.47 | -3.64 | -3.59 |
| Average | -2.26 | 4.85 | -1.38 | -0.46 | -3.93 | -4.22 | -0.92 | -3.09 | -0.96 | -2.80 | -1.76 |
Table 1 Energy decomposition of the binding affinity between the amino acid(AA) residues and phenolic substrates
| Substrate | ΔEb/(kJ·mol-1) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Leu164 | Asp206 | Asn208 | Phe239 | Asn264 | Phe265 | Pro391 | Gly392 | Ala393 | Pro394 | Hie458 | |
| 1 | -1.34 | 9.45 | -1.34 | -0.42 | -3.51 | -3.55 | -0.29 | -2.80 | 0 | -2.76 | -1.00 |
| 2 | -2.42 | -1.42 | -0.59 | -0.29 | -1.67 | -4.05 | -0.38 | -2.13 | -0.79 | -1.80 | -1.76 |
| 3 | -0.84 | -8.99 | -1.96 | -0.67 | -3.64 | -3.30 | -0.25 | -0.84 | -1.42 | -1.21 | -1.55 |
| 4 | -1.09 | -2.26 | -0.96 | -0.42 | -2.55 | -3.64 | -0.29 | -1.38 | -0.67 | -1.88 | -1.05 |
| 5 | -0.75 | 9.15 | -2.80 | -0.67 | -5.27 | -3.34 | -0.25 | -1.21 | -1.00 | -2.22 | -0.79 |
| 6 | -0.84 | -4.39 | -4.68 | -0.96 | -6.73 | -4.39 | -0.25 | -0.96 | -0.42 | -2.72 | -2.09 |
| 7 | -2.63 | 2.30 | -0.50 | -0.33 | -0.84 | -4.05 | -1.05 | -2.34 | -0.92 | -2.22 | -2.68 |
| 8 | -1.46 | 6.44 | -2.01 | -0.50 | -5.73 | -4.56 | -0.25 | -3.39 | -0.67 | -3.97 | -0.75 |
| 9 | -2.13 | 9.78 | -0.96 | -0.50 | -5.35 | -3.01 | -0.38 | -3.55 | -0.38 | -2.51 | -0.54 |
| 10 | -1.71 | 8.40 | -0.84 | -0.46 | -6.10 | -3.51 | -0.33 | -3.55 | -2.97 | -2.93 | -0.71 |
| 11 | -1.34 | 6.77 | -2.05 | -0.46 | -6.10 | -4.35 | -0.25 | -2.88 | -1.25 | -4.22 | -1.09 |
| 12 | -2.63 | 7.15 | -2.05 | -0.54 | -3.89 | -3.68 | -0.46 | -4.39 | -0.29 | -2.88 | -0.88 |
| 13 | -2.26 | 9.24 | -0.92 | -0.42 | -4.47 | -3.14 | -0.50 | -4.47 | 0.04 | -1.71 | -0.71 |
| 14 | -2.34 | 3.76 | -1.09 | -0.46 | -1.25 | -4.60 | -0.42 | -2.63 | -0.42 | -2.55 | -1.30 |
| 15 | -2.51 | -6.94 | -0.75 | -0.42 | -1.59 | -3.55 | -0.46 | -2.88 | -0.59 | -1.80 | -1.00 |
| 16 | -1.30 | 8.74 | -1.76 | -0.46 | -6.23 | -4.01 | -0.33 | -2.84 | -0.04 | -3.05 | -0.88 |
| 17 | -1.50 | 8.28 | -1.00 | -0.46 | -4.72 | -4.85 | -0.25 | -2.30 | -1.30 | -3.55 | -0.88 |
| 18 | -2.30 | 10.24 | -0.88 | -0.46 | -2.93 | -6.60 | -5.77 | -5.64 | -3.68 | -3.59 | -5.98 |
| 19 | -8.53 | 8.40 | -0.13 | -0.21 | 0.04 | -3.05 | -4.81 | -6.60 | 1.34 | -1.67 | -4.39 |
| 20 | -4.97 | 8.07 | -0.75 | -0.38 | -4.18 | -5.31 | -0.92 | -4.93 | -1.38 | -5.94 | -3.72 |
| 21 | -2.68 | 9.91 | -1.38 | -0.42 | -5.31 | -7.77 | -1.21 | -3.68 | -3.47 | -3.64 | -3.59 |
| Average | -2.26 | 4.85 | -1.38 | -0.46 | -3.93 | -4.22 | -0.92 | -3.09 | -0.96 | -2.80 | -1.76 |
| Position | Model | R(2-ortho) | R(6-ortho) | R(para) | ΔΔG/(kJ·mol-1) | ΔΔG'/(kJ·mol-1) | Taft σ* | σpara |
|---|---|---|---|---|---|---|---|---|
| 2-ortho- | 4 | NH2 | H | H | 0.00 | 0.62 | ||
| 3 | OH | H | H | 39.33 | 1.34 | |||
| 5 | OCH3 | H | H | 40.84 | 1.81 | |||
| 7 | CH=CHCH3 | H | H | 43.72 | 0.36 | |||
| 1 | H | H | H | 55.76 | 0.49 | |||
| 2 | Cl | H | H | 78.50 | 2.96 | |||
| 6 | CONH2 | H | H | 98.69 | 1.68 | |||
| ortho, | 9 | OCH3 | H | NH2 | -8.78 | -49.62a | -0.66 | |
| para- | 10 | OCH3 | H | OH | 19.31 | -21.53a | -0.37 | |
| 8 | OCH3 | H | Cl | 46.44 | 5.60a | 0.23 | ||
| 11 | OCH3 | H | CONH2 | 53.88 | 13.04a | 0.36 | ||
| 2,6- | 12 | OCH3 | OCH3 | H | -19.77 | -60.61a | 3.62 | |
| 13 | OCH3 | OH | H | -0.84 | -41.67a | 3.15 | ||
| 15 | OH | OH | H | 45.48 | 6.14b | 2.68 | ||
| 14 | Cl | Cl | H | 76.37 | -2.17c | 5.92 | ||
| Lignin | 21 | OCH3 | H | CHR''OH | 11.75 | |||
| model | 18 | OCH3 | PhR | CH2OH | 25.50 | |||
| compounds | 20 | OCH3 | H | CHR'OH | 37.91 | |||
| 16 | OCH3 | H | CH2OH | 42.85 | ||||
| 17 | OCH3 | H | CHO | 72.31 | ||||
| 19 | OCH3 | H | COR' | 76.95 |
Table 2 Comparison of Gibbs free energy changes among different phenolic substrates
| Position | Model | R(2-ortho) | R(6-ortho) | R(para) | ΔΔG/(kJ·mol-1) | ΔΔG'/(kJ·mol-1) | Taft σ* | σpara |
|---|---|---|---|---|---|---|---|---|
| 2-ortho- | 4 | NH2 | H | H | 0.00 | 0.62 | ||
| 3 | OH | H | H | 39.33 | 1.34 | |||
| 5 | OCH3 | H | H | 40.84 | 1.81 | |||
| 7 | CH=CHCH3 | H | H | 43.72 | 0.36 | |||
| 1 | H | H | H | 55.76 | 0.49 | |||
| 2 | Cl | H | H | 78.50 | 2.96 | |||
| 6 | CONH2 | H | H | 98.69 | 1.68 | |||
| ortho, | 9 | OCH3 | H | NH2 | -8.78 | -49.62a | -0.66 | |
| para- | 10 | OCH3 | H | OH | 19.31 | -21.53a | -0.37 | |
| 8 | OCH3 | H | Cl | 46.44 | 5.60a | 0.23 | ||
| 11 | OCH3 | H | CONH2 | 53.88 | 13.04a | 0.36 | ||
| 2,6- | 12 | OCH3 | OCH3 | H | -19.77 | -60.61a | 3.62 | |
| 13 | OCH3 | OH | H | -0.84 | -41.67a | 3.15 | ||
| 15 | OH | OH | H | 45.48 | 6.14b | 2.68 | ||
| 14 | Cl | Cl | H | 76.37 | -2.17c | 5.92 | ||
| Lignin | 21 | OCH3 | H | CHR''OH | 11.75 | |||
| model | 18 | OCH3 | PhR | CH2OH | 25.50 | |||
| compounds | 20 | OCH3 | H | CHR'OH | 37.91 | |||
| 16 | OCH3 | H | CH2OH | 42.85 | ||||
| 17 | OCH3 | H | CHO | 72.31 | ||||
| 19 | OCH3 | H | COR' | 76.95 |
| [1] | Mayer A. M., Staples R. C., Phytochemistry,2002, 60(6), 551—565 |
| [2] | Bao W., O'malley D. M., Whetten R., Sederoff R. R., Science,1993, 260(5108), 672 |
| [3] | Wan Y. Y., Miyakoshi T., Du Y. M., Chen L. J., Hao J. M., Kennedy J. F., Int. J. Biol. Macromol., 2012, 50(3), 530—533 |
| [4] | Giardina P., Faraco V., Pezzella C., Piscitelli A., Vanhulle S., Sannia G., Cellular and Molecular Life Sciences,2010, 67(3), 369—385 |
| [5] | Dai Y., Yin L., Niu J., Environmental Science & Technology,2011, 45(24), 10611—10618 |
| [6] | Lu J., Huang Q., Mao L., Environmental Science & Technology,2009, 43(18), 7062—7067 |
| [7] | Feng Y., Colosi L. M., Gao S., Huang Q., Mao L., Environmental Science & Technology,2012, 47(2), 1001—1008 |
| [8] | Baldrian P., FEMS Microbiology Reviews, 2006, 30(2), 215—242 |
| [9] | Solomon E. I., Sundaram U. M., Machonkin T. E., Chemical Reviews,1996, 96(7), 2563—2606 |
| [10] | Piontek K., Antorini M., Choinowski T., J. Biol. Chem., 2002, 277(40), 37663—37669 |
| [11] | Jeon J. R., Chang Y. S., TRENDS in Biotechnology,2013, 31(6), 335—341 |
| [12] | Prasad N. K., Vindal V., Narayana S. L., Kunal S. P., J. Mole. Mode., 2011, 18(5), 2013—2019 |
| [13] | Cambria M. T., Di Marino D., Falconi M., Garavaglia S., Cambria A., J. Biomol. Struct. Dyn., 2010, 27(4), 501—509 |
| [14] | Galli C., Gentili P., Jolivalt C., Madzak C., Vadalà R., Applied Microbiology and Biotechnology,2011, 91(1), 123—131 |
| [15] | Schomburg I., Chang A., Placzek S., Söhngen C., Rother M., Lang M., Munaretto C., Ulas S., Stelzer M., Grote A., Nucleic Acids Research,2013, 41(D1), D764—D772 |
| [16] | Eggert C., Temp U., Dean J. F., Eriksson K. E. L., FEBS Letters,1996, 391(1), 144—148 |
| [17] | Lahtinen M., Kruus K., Boer H., Kemell M., Andberg M., Viikari L., Sipilä J., J. Mole. Cata. B: Enzymatic,2009, 57(1), 204—210 |
| [18] | The PyMOL Molecular Graphics System, Version 1.3r1, Schrödinger LLC, Version 1.3r1, Schrödinger LLC, Portland OR, 2010 |
| [19] | Rastelli G., Rio A. D., Degliesposti G., Sgobba M., J. Comput. Chem., 2010, 31(4), 797—810 |
| [20] | Case D., Darden T., Cheatham III T., Simmerling C., Wang J., Duke R., Luo R., Walker R., Zhang W., Merz K., Amber, Version 10, University of California, San Francisco, 2012 |
| [21] | Mahoney M. W., Jorgensen W. L., J. Chem. Phys., 2000, 112(20), 8910—8922 |
| [22] | Miller B. R. III, McGee T. D. Jr., Swails J. M., Homeyer N., Gohlke H., Roitberg A. E., J. Chem. Theo. Comput., 2012, 8(9), 3314—3321 |
| [23] | Bertrand T., Jolivalt C., Briozzo P., Caminade E., Joly N., Madzak C., Mougin C., Biochemistry,2002, 41(23), 7325—7333 |
| [24] | Frisch M.J., Trucks G. W., Schlegel H. B., Scuseria G. E., Robb M. A., Cheeseman J. R., Scalmani G., Barone V., Mennucci B., Petersson G. A., Nakatsuji H., Caricato M., Li X., Hratchian H. P., Izmaylov A. F., Bloino J., Zheng G., Sonnenberg J. L., Hada M., Ehara M., Toyota K., Fukuda R., Hasegawa J., Ishida M., Nakajima T., Honda Y., Kitao O., Nakai H., Vreven T., Montgomery J. A. Jr., Peralta J. E., Ogliaro F., Bearpark M., Heyd J. J., Brothers E., Kudin K. N., Staroverov V. N., Kobayashi R., Normand J., Raghavachari K., Rendell A., Burant J. C., Iyengar S. S., Tomasi J., Cossi M., Rega N., Millam N. J., Klene M., Knox J. E., Cross J. B., Bakken V., Adamo C., Jaramillo J., Gomperts R., Stratmann R. E., Yazyev O., Austin A. J., Cammi R., Pomelli C., Ochterski J. W., Martin R. L., Morokuma K., Zakrzewski V. G., Voth G. A., Salvador P., Dannenberg J. J., Dapprich S., Daniels A. D., Farkas Ö., Foresman J. B., Ortiz J. V., Cioslowski J., Fox D. J., Gaussian 09, Revision A.02, Gaussian Inc., Wallingford CT, 2009 |
| [25] | Becke A. D., J. Chem. Phys., 1993, 98(2), 1372—1377 |
| [26] | Kallio J., Auer S., Janis J., Andberg M., Kruus K., Rouvinen J., Koivula A., Hakulinen N., J. Mole. Bio., 2009, 392(4), 895—909 |
| [27] | Tadesse M. A., D’Annibale A., Galli C., Gentili P., Sergi F., Org. Biomol. Chem., 2008, 6(5), 868—878 |
| [28] | Dean J.A., Lange’s Handbook of Chemistry, McGraw-Hill Inc., New York, 1985, 1348—1352 |
| [29] | Galli C., Madzak C., Vadalà R., Jolivalt C., Gentili P., Chem. Bio. Chem., 2013, 14(8), 2500—2505 |
| [30] | Weinberg D. R., Gagliardi C. J., Hull J. F., Murphy C. F., Kent C. A., Westlake B. C., Paul A., Ess D. H., McCafferty D. G., Meyer T. J., Chemical Reviews,2012, 112(7), 4016—4093 |
| [31] | Hammes-Schiffer S., Proceedings of the National Academy of Sciences, 2011, 108(21), 8531—8532 |
| [32] | Madzak C., Mimmi M., Caminade E., Brault A., Baumberger S., Briozzo P., Mougin C., Jolivalt C., Protein Engineering Design and Selection,2006, 19(2), 77—84 |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||