

高等学校化学学报 ›› 2024, Vol. 45 ›› Issue (11): 20240355.doi: 10.7503/cjcu20240355
黄玉滢1,2, 于成鲲1,2, 刘斯奇2,3, 任艳2,4( )
)
收稿日期:2024-07-15
出版日期:2024-11-10
发布日期:2024-09-02
通讯作者:
任艳
E-mail:reny@genomics.cn
基金资助:
HUANG Yuying1,2, YU Chengkun1,2, LIU Siqi2,3, REN Yan2,4( )
)
Received:2024-07-15
Online:2024-11-10
Published:2024-09-02
Contact:
REN Yan
E-mail:reny@genomics.cn
Supported by:摘要:
采用抗体荧光标记法分离出T细胞、 B细胞、 自然杀伤细胞和树突状细胞. 使用CellenONE单细胞分选系统分选出相应的单细胞, 在分选过程中应用了质谱兼容的肽段包被单细胞蛋白质组学(Mad-CASP)技术. 将高疏水性肽段预先加入至分选的孔板中, 从而减少了蛋白在孔板和色谱柱上的吸附. 提取出单细胞蛋白并进行酶解, 采用液相色谱-质谱联用(LC-MS/MS)技术对获得的肽段进行数据采集, 并利用Maxquant软件中的谱图库及“运行中匹配”功能进行了蛋白质的搜库鉴定. 采用维恩图和统一流形近似与投影(UMAP)技术分析了 4种细胞的蛋白表达差异, 对细胞的特异性蛋白进行了分子特征数据库小鼠免疫通路富集, 并对计数排序前2名的通路进行了分析, 同时利用模糊C均值聚类方法和京都大学基因与基因组数据库(KEGG)通路富集分析了免疫细胞共享蛋白定量值的变化, 绘制了专属于小鼠外周血单个核细胞的单细胞蛋白质组学图谱. 研究结果对深入理解免疫细胞的功能特征和发现疾病相关的关键蛋白标记物具有重要价值.
中图分类号:
TrendMD:
黄玉滢, 于成鲲, 刘斯奇, 任艳. 利用无标记单细胞蛋白质组学方法构建小鼠外周血单个核细胞的细胞图谱. 高等学校化学学报, 2024, 45(11): 20240355.
HUANG Yuying, YU Chengkun, LIU Siqi, REN Yan. Cell Map of Mouse Peripheral Blood Mononuclear Cells with a Label-free Single-cell Proteomics Method. Chem. J. Chinese Universities, 2024, 45(11): 20240355.
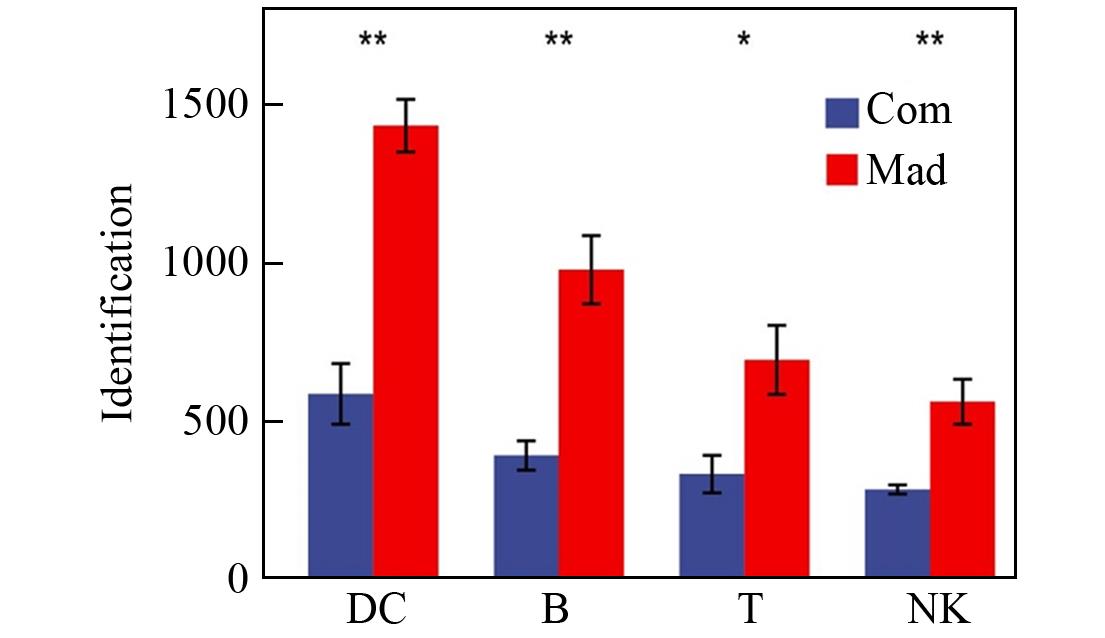
Fig.3 Comparison of the number of protein identifications between the common pretreatment technique and the Mad⁃CASP techniqueThe error bars represent the standard deviation.* indicates P<0.05, ** indicates P<0.01, n=5.
| 1 | Teo Y. V., Hinthorn S. J., Webb A. E., Neretti N., Aging, 2023, 15(1), 6—20 |
| 2 | Kaech S. M., Wherry E. J., Ahmed R., Nat. Rev. Immunol., 2002, 2(4), 251—262 |
| 3 | Liu Z., Li J. P., Chen M., Wu M., Shi Y., Li W., Teijaro J. R., Wu P., Cell, 2020, 183(4), 1117—1133.e19 |
| 4 | Sakaguchi S., Yamaguchi T., Nomura T., Ono M., Cell, 2008, 133(5), 775—787 |
| 5 | Kurosaki T., Kometani K., Ise W., Nat. Rev. Immunol., 2015, 15(3), 149—159 |
| 6 | Liu Y., Beyer A., Aebersold R., Cell, 2016, 165(3), 535—550 |
| 7 | Segel M., Neumann B., Hill M. F. E., Weber I. P., Viscomi C., Zhao C., Young A., Agley C. C., Thompson A. J., Gonzalez G. A., Sharma A., Holmqvist S., Rowitch D. H., Franze K., Franklin R. J. M., Chalut K. J., Nature, 2019, 573(7772), 130—134 |
| 8 | Specht H., Slavov N., J. Proteome Res., 2018, 17(8), 2565—2571 |
| 9 | Levy E., Slavov N., Essays Biochem., 2018, 62(4), 595—605 |
| 10 | Spitzer M. H., Nolan G. P., Cell, 2016, 165(4), 780—791 |
| 11 | Lombard⁃Banek C., Reddy S., Moody S. A., Nemes P., Mol. Cell. Proteomics, 2016, 15(8), 2756—2768 |
| 12 | Petelski A. A., Emmott E., Leduc A., Huffman R. G., Specht H., Perlman D. H., Slavov N., Nat. Protoc., 2021, 16(12), 5398—5425 |
| 13 | Thielert M., Itang E. C., Ammar C., Rosenberger F. A., Bludau I., Schweizer L., Nordmann T. M., Skowronek P., Wahle M., Zeng W. F., Zhou X. X., Brunner A. D., Richter S., Levesque M. P., Theis F. J., Steger M., Mann M., Mol. Syst. Biol., 2023, 19(9), e11503 |
| 14 | Zhu Y., Piehowski P. D., Zhao R., Chen J., Shen Y., Moore R. J., Shukla A. K., Petyuk V. A., Campbell⁃Thompson M., Mathews C. E., Smith R. D., Qian W. J., Kelly R. T., Nat. Commun., 2018, 9(1), 882 |
| 15 | Ctortecka C., Clark N. M., Boyle B. W., Seth A., Mani D. R., Udeshi N. D., Carr S. A., Nat. Commun., 2024, 15(1), 5707 |
| 16 | Budnik B., Levy E., Harmange G., Slavov N., Genome Biol., 2018, 19(1), 161 |
| 17 | Rosenberger F. A., Thielert M., Mann M., Nat. Methods, 2023, 20(3), 320—323 |
| 18 | Matzinger M., Müller E., Dürnberger G., Pichler P., Mechtler K., Anal. Chem., 2023, 95(9), 4435—4445 |
| 19 | Li S., Su K., Zhuang Z., Qin Q., Gao L., Deng Y., Liu X., Hou G., Wang L., Hao P., Yang H., Liu S., Zhu H., Ren Y., Sci. Bull., 2022, 67(6), 581—584 |
| 20 | Kebarle P., Verkerk U. H., Mass Spectrom. Rev., 2009, 28(6), 898—917 |
| 21 | Cox J., Mann M., Nat. Biotechnol., 2008, 26(12), 1367—1372 |
| 22 | Cox J., Hein M. Y., Luber C. A., Paron I., Nagaraj N., Mann M., Mol. Cell. Proteomics, 2014, 13(9), 2513—2526 |
| 23 | Becht E., McInnes L., Healy J., Dutertre C. A., Kwok I. W. H., Ng L. G., Ginhoux F., Newell E. W., Nat. Biotechnol., 2019, 37(1), 38—44 |
| 24 | Liberzon A., Subramanian A., Pinchback R., Thorvaldsdóttir H., Tamayo P., Mesirov J. P., Bioinformatics 2011, 27(12), 1739—1740 |
| 25 | Ferri F., Parcelier A., Petit V., Gallouet A. S., Lewandowski D., Dalloz M., van den Heuvel A., Kolovos P., Soler E., Squadrito M. L., De Palma M., Davidson I., Rousselet G., Romeo P. H., Nat. Commun., 2015, 6, 8900 |
| 26 | Banchereau J., Steinman R. M., Nature, 1998, 392(6673), 245—252 |
| 27 | Deng Y., Xie M., Li Q., Xu X., Ou W., Zhang Y., Xiao H., Yu H., Zheng Y., Liang Y., Jiang C., Chen G., Du D., Zheng W., Wang S., Gong M., Chen Y., Tian R., Li T., Circ. Res., 2021, 128(2), 232—245 |
| 28 | Kumar L., Futschik M. E., Bioinformation, 2007, 2(1), 5—7 |
| 29 | Bezdek J. C., Pattern Recognition with Fuzzy Objective Function Algorithms, Plenum Press, New York, 1981, 191—226 |
| [1] | 毕英娜 刘定斌. 基于内标法的表面增强拉曼光谱在定量分析中的应用[J]. 高等学校化学学报, 0, (): 20240457. |
| [2] | 董沛滢, 刘彤, 秦伟捷. RNA-蛋白质复合物规模化富集与鉴定新方法[J]. 高等学校化学学报, 2024, 45(11): 20240091. |
| [3] | 汪增钰, 刘宝红, 乔亮, 林灵. 非靶向脂质组学揭示巨噬细胞泡沫化进程脂质代谢功能失调[J]. 高等学校化学学报, 2024, 45(11): 20240053. |
| [4] | 左春倩, 许瑞瑞, 毕红燕. 水产品中过敏原检测的研究进展[J]. 高等学校化学学报, 2024, 45(11): 20240073. |
| [5] | 陈天泽, 胡方舟, 林绪波, 应佚伦, 邹爱华. 基于Aerolysin纳米孔道的单个β-淀粉样多肽N-端片段分析[J]. 高等学校化学学报, 2024, 45(11): 20240192. |
| [6] | 石倩, 刘冬梅, 方小泥, 刘宝红. 基于基质辅助激光解析质谱的高通量酪氨酸酶活性及抑制剂分析[J]. 高等学校化学学报, 2024, 45(11): 20240330. |
| [7] | 蒋龑, 陈妍琳, 宋高瑜, 陈炎炎, 白晶, 朱莹娣, 李娟. 细菌的蛋白质组成分析[J]. 高等学校化学学报, 2024, 45(11): 20240345. |
| [8] | 闫勇杰, 高文博, 鲁晨辉, 杨成, 徐姝婷. 基于微萃取-纳喷雾质谱技术的纳升脑脊液中咖啡多酚的检测[J]. 高等学校化学学报, 2024, 45(11): 20240327. |
| [9] | 靳莹, 张俊杰, 张毅欣, 袁悦, 韩珍珍. 外泌体分离和蛋白质组学分析的研究进展[J]. 高等学校化学学报, 2024, 45(11): 20240305. |
| [10] | 胡宇虹, 俞相明, 宋丽丽, 邢清和, 周峰. DEEP SEQ方法检测新生儿毛干蛋白质组动态变化[J]. 高等学校化学学报, 2024, 45(11): 20240326. |
| [11] | 范智瑞, 方群, 杨奕. 基于质谱的单细胞蛋白质组学分析[J]. 高等学校化学学报, 2024, 45(11): 20240294. |
| [12] | 张磊, 申华莉. 标志物研究中常用蛋白组学质谱方法的定量准确性评估[J]. 高等学校化学学报, 2024, 45(11): 20240311. |
| [13] | 续红妹, 王梁臣, 闵乾昊. 面向小分子检测的MALDI MS基质研究进展[J]. 高等学校化学学报, 2024, 45(11): 20240285. |
| [14] | 刘思盈, 粟雯, 周仲燕, 杨治渝, 裴华夫, 何芷茹, 王娜, 岳磊. 温度控制的泛素/三磷酸腺苷相互作用的电喷雾质谱研究[J]. 高等学校化学学报, 2024, 45(11): 20240382. |
| [15] | 沈枫林, 冯兆莹, 方静, 张磊, 刘晓慧, 周新文. 基于质谱的单细胞分辨的空间蛋白质组学新技术研究[J]. 高等学校化学学报, 2024, 45(11): 20240299. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||