

高等学校化学学报 ›› 2024, Vol. 45 ›› Issue (11): 20240389.doi: 10.7503/cjcu20240389
• 综合评述 • 上一篇
霍志远1,2, 周金萍3, 马秀敏3( ), 周严1,2(
), 周严1,2( ), 黄琳1,2(
), 黄琳1,2( )
)
收稿日期:2024-08-15
出版日期:2024-11-10
发布日期:2024-09-11
通讯作者:
马秀敏,周严,黄琳
E-mail:maxiumin1210@sohu.com;zhouyan@shchest.org;linhuang@shsmu.edu.cn
作者简介:第一联系人:共同第一作者.
基金资助:
HUO Zhiyuan1,2, ZHOU Jinping3, MA Xiumin3( ), ZHOU Yan1,2(
), ZHOU Yan1,2( ), HUANG Lin1,2(
), HUANG Lin1,2( )
)
Received:2024-08-15
Online:2024-11-10
Published:2024-09-11
Contact:
MA Xiumin, ZHOU Yan, HUANG Lin
E-mail:maxiumin1210@sohu.com;zhouyan@shchest.org;linhuang@shsmu.edu.cn
Supported by:摘要:
近年来, 随着质谱技术的飞速发展, 单细胞质谱技术在生命科学领域发挥着越来越重要的作用. 质谱已被广泛用于检测细胞群代谢和蛋白组, 单细胞质谱多组学技术结合微观生物学和尖端质谱分析, 为在单细胞层面进行深入研究提供了强有力的工具, 进一步阐明了细胞微观复杂性. 特别是在生物医学应用中, 单细胞质谱多组学有助于细胞图谱绘制、 生命现象分子机制探究及精准医学的发展等. 本文综合评述了近年来基于质谱的单细胞代谢、 蛋白组及其整合组学技术研发中面临的挑战与突破, 并对其未来的发展趋势进行了展望.
中图分类号:
TrendMD:
霍志远, 周金萍, 马秀敏, 周严, 黄琳. 基于质谱的单细胞多组学分析技术研究进展. 高等学校化学学报, 2024, 45(11): 20240389.
HUO Zhiyuan, ZHOU Jinping, MA Xiumin, ZHOU Yan, HUANG Lin. Advances in Single-cell Multi-omics Analysis Based on Mass Spectrometry. Chem. J. Chinese Universities, 2024, 45(11): 20240389.
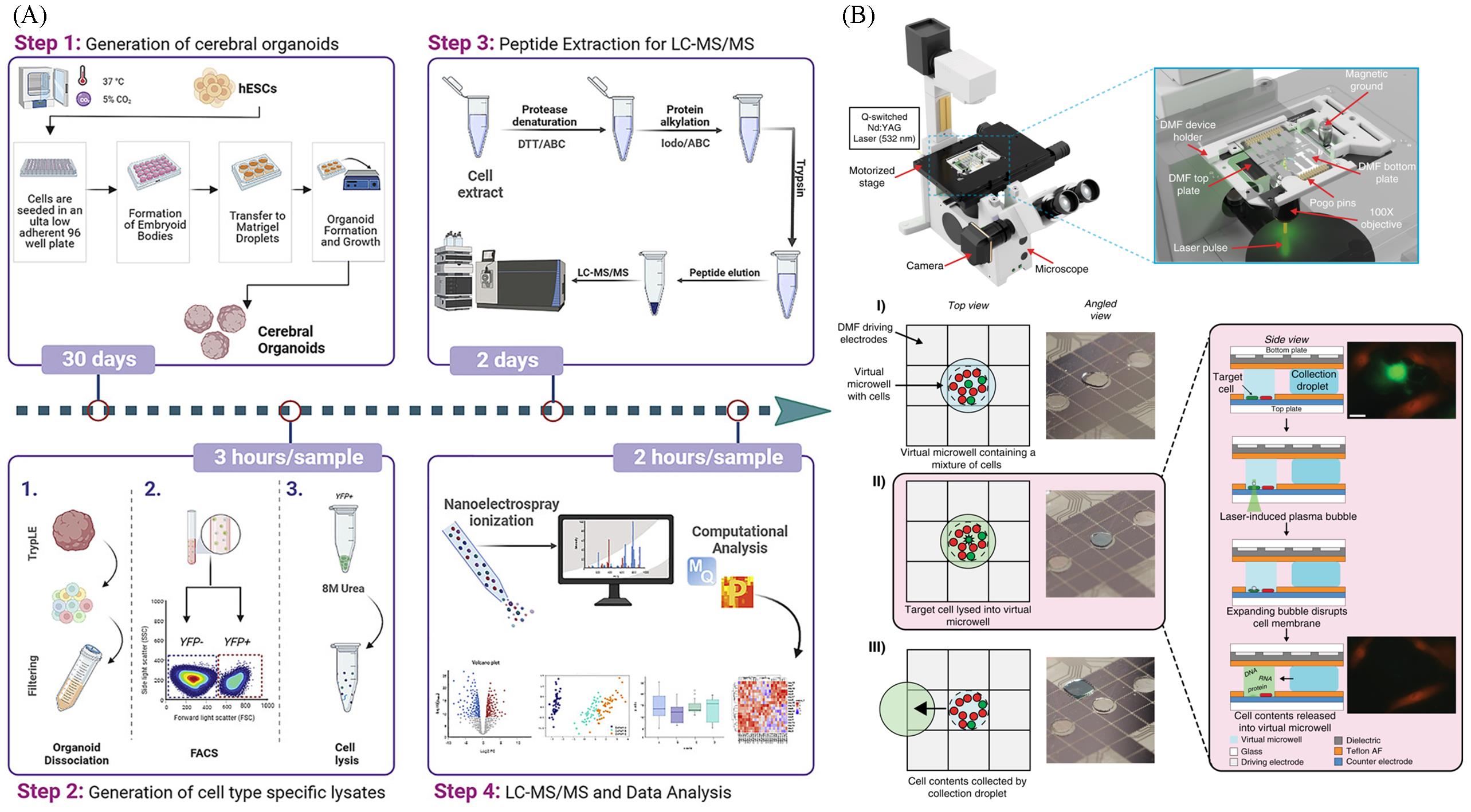
Fig.1 Research progress in single⁃cell separation and capture technologies(A) FACS for single-cell proteomics[12]. Copyright 2022, Elsevier; (B) digital microfluidic technique for single-cell proteomics [18]. Copyright 2020, Springer Nature.
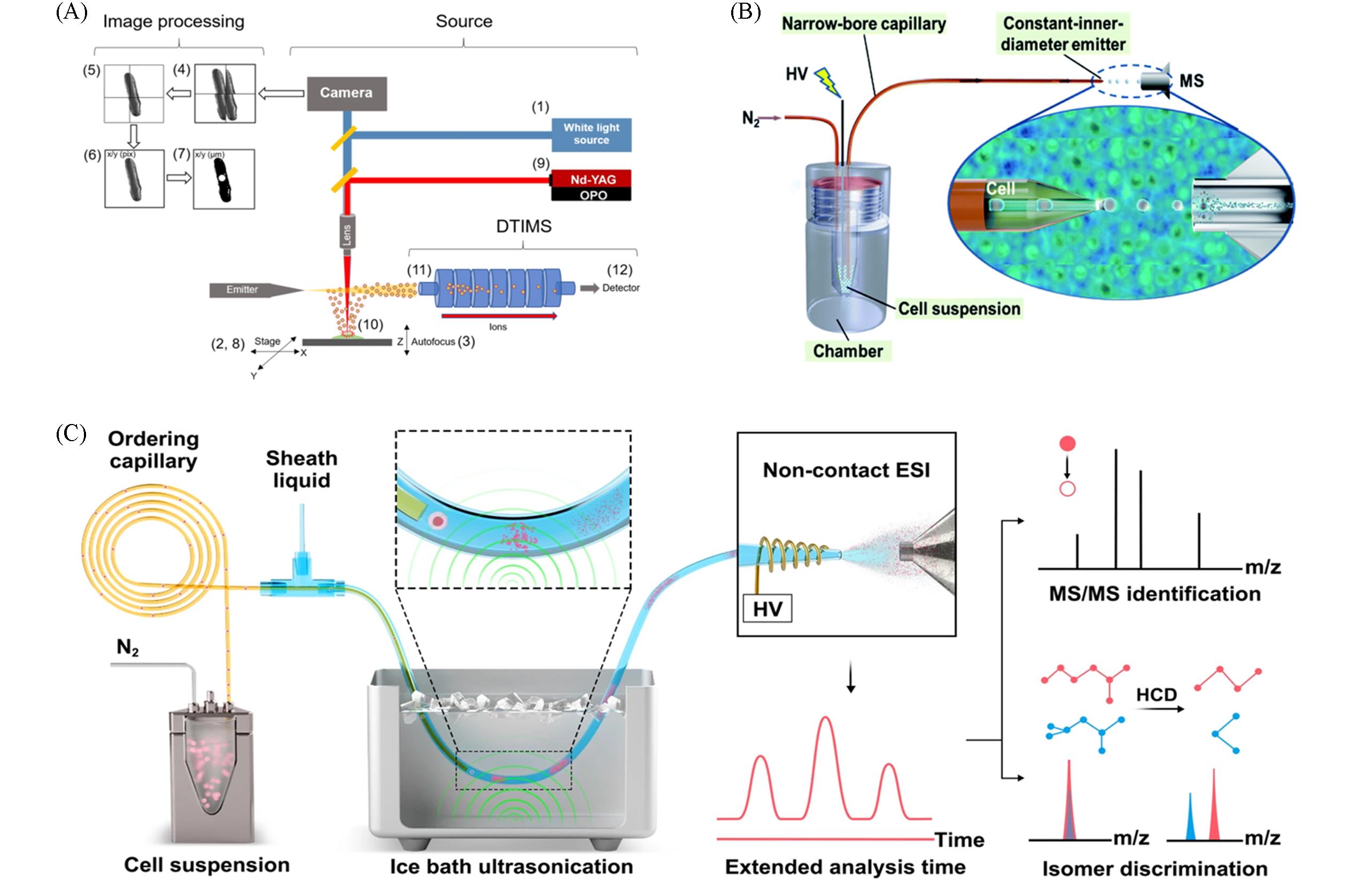
Fig.2 Research progress in single⁃cell mass spectrometry metabolomics(A) Schematic of LAESI-DTIMS based on microscope system[28]. Copyright 2021, Molecular Diversity Preservation International; (B) schematic illustration of the ILCEI-MS system[27]. Copyright 2022, the Royal Society of Chemistry; (C) the workflow of ID-organic cytoMS[26]. Copyright 2024, Springer Nature.
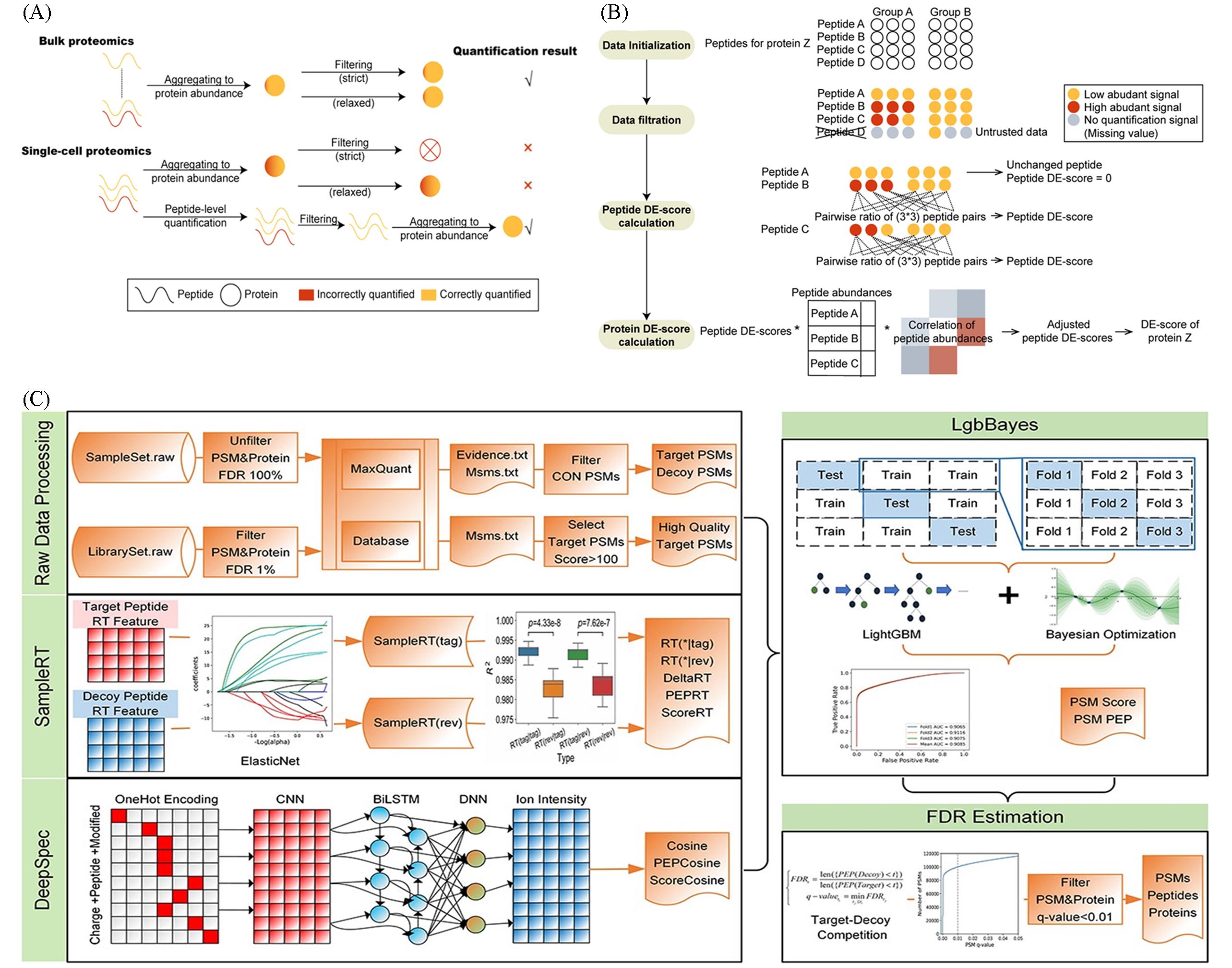
Fig.3 Research progress in single⁃cell mass spectrometry proteomics(A, B) The workflow of pepDESC is illustrated by protein Z[36]. Copyright 2023, Elsevier; (C) the overall framework of DeepSCP[35]. Copyright 2022, Oxford University Press.
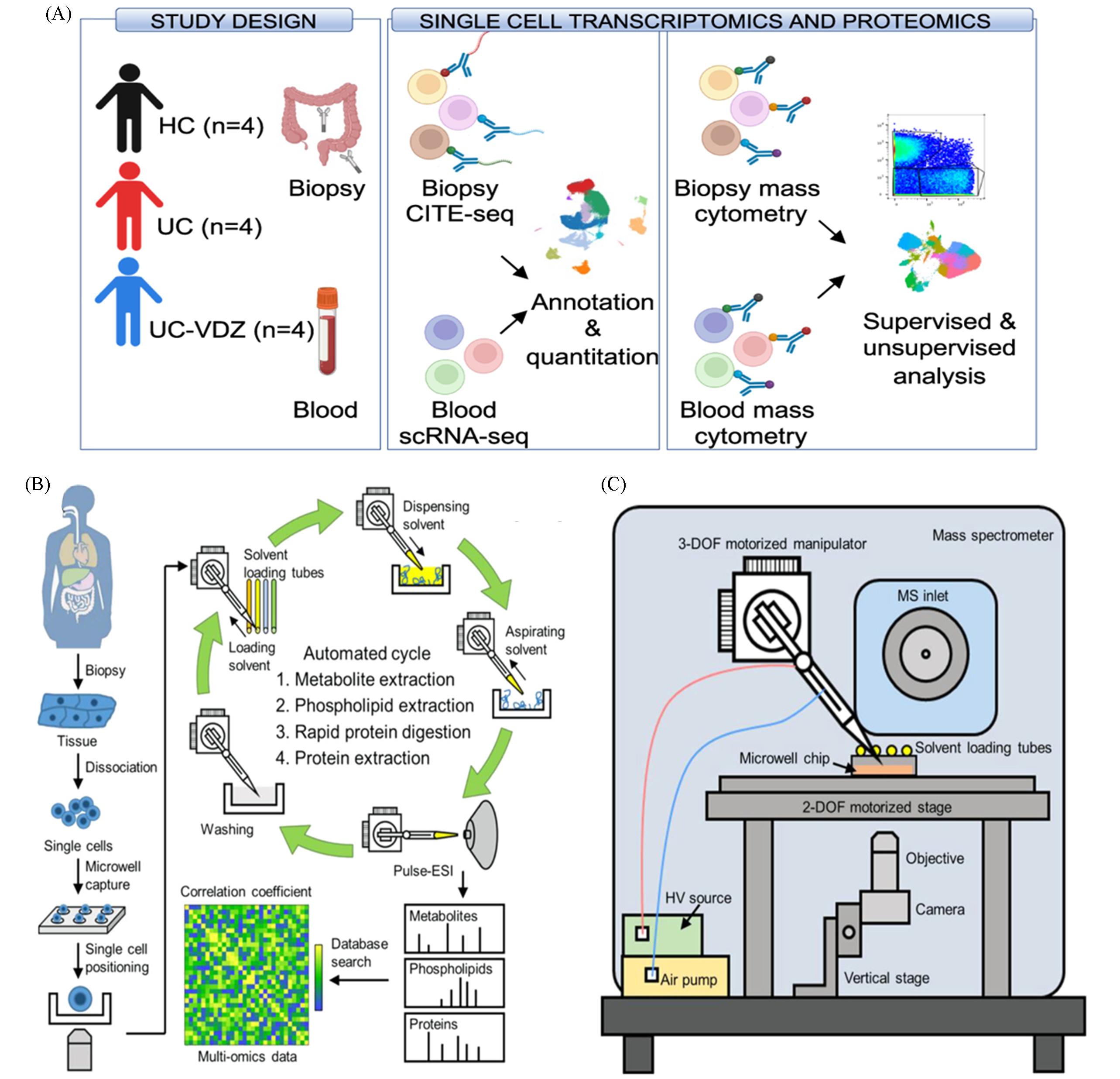
Fig.4 Research progress in single⁃cell mass spectrometry multi⁃omics(A) Schematic of platform design combining single-cell transcriptomics(CITE-seq) and proteomics(CyTOF)[45]. Copyright 2024, Springer Nature; (B, C) schematic of the highly efficient and automatic single-cell multi-omics mass spectrometry strategy[47]. Copyright 2023, American Chemical Society.
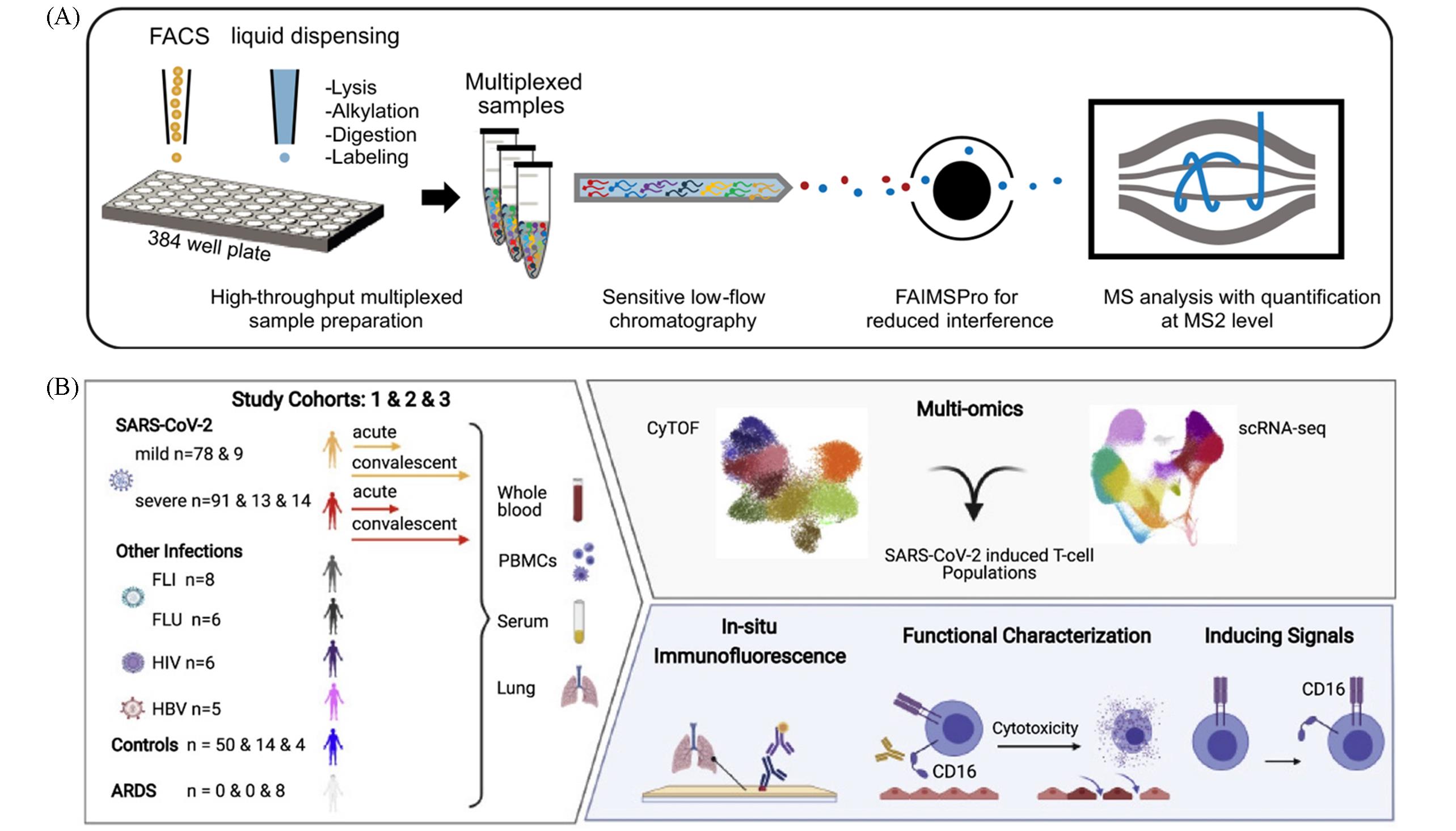
Fig.5 Biomedical applications of single⁃cell mass spectrometry multi⁃omics(A) Quantitative LC-MS-based single-cell proteomics(scMS) serves as a tool for characterizing cellular hierarchies[51]. Copyright 2021, Springer Nature; (B) platform combining single-cell transcriptomics and single-cell proteomics for biomarker discovery in severe COVID-19[55]. Copyright 2022, Elsevier.
| 1 | Baysoy A., Bai Z., Satija R., Fan R., Nat. Rev. Mol. Cell Biol., 2023, 24(10), 695—713 |
| 2 | Flynn E., Almonte⁃Loya A., Fragiadakis G. K., Annu. Rev. Biomed., 2023, 6(1), 313—337 |
| 3 | Macaulay I. C., Ponting C. P., Voet T., Trends Genet., 2017, 33(2), 155—168 |
| 4 | Zhu Q., Zhao X., Zhang Y., Li Y., Liu S., Han J., Sun Z., Wang C., Deng D., Wang S., Nat. Commun., 2023, 14(1), 8170 |
| 5 | Ma A., McDermaid A., Xu J., Chang Y., Ma Q., Trends Biotechnol., 2020, 38(9), 1007—1022 |
| 6 | Chen W., Yu H., Hao Y., Liu W., Wang R., Huang Y., Wu J., Feng L., Guan Y., Huang L., ACS Nano, 2023, 17(20), 19779—19792 |
| 7 | Wang Y., Xu X., Fang Y., Yang S., Wang Q., Liu W., Zhang J., Liang D., Zhai W., Qian K., ACS Nano, 2024, 18(3), 2409— 2420 |
| 8 | Wu J., Wei X., Gan J., Huang L., Shen T., Lou J., Liu B., Zhang J. X., Qian K., Adv. Funct. Mater., 2016, 26(22), 4016—4025 |
| 9 | Zhang J., Teng F., Hu B., Liu W., Huang Y., Wu J., Wang Y., Su H., Yang S., Zhang L., Adv. Mater., 2024, 36(18), 2311431 |
| 10 | Herzenberg L. A., Parks D., Sahaf B., Perez O., Roederer M., Herzenberg L. A., Clinical Chem., 2002, 48(10), 1819—1827 |
| 11 | Sherman S. E., Kuljanin M., Cooper T. T., Lajoie G. A., Hess D. A., Stem Cells Dev., 2020, 29(14), 895—910 |
| 12 | Melliou S., Diamandis P., Star Protoc., 2022, 3(4), 101774 |
| 13 | Espina V., Heiby M., Pierobon M., Liotta L. A., Expert Rev. Mol. Diagn., 2007, 7(5), 647—657 |
| 14 | Guise A. J., Misal S. A., Carson R., Chu J. H., Boekweg H., van Der Watt D., Welsh N. C., Truong T., Liang Y., Xu S., Benedetto G., Gagnon J., Payne S. H., Plowey E. D., Kelly R. T., Cell Rep., 2024, 43(1), 113636 |
| 15 | Rosenberger F. A., Thielert M., Strauss M. T., Schweizer L., Ammar C., Mädler S. C., Metousis A., Skowronek P., Wahle M., Madden K., Gote⁃Schniering J., Semenova A., Schiller H. B., Rodriguez E., Nordmann T. M., Mund A., Mann M., Nat. Methods, 2023, 20(10), 1530—1536 |
| 16 | Lecault V., White A. K., Singhal A., Hansen C. L., Curr. Opin. Chem. Biol., 2012, 16(3/4), 381—390 |
| 17 | Feng D., Li H., Xu T., Zheng F., Hu C., Shi X., Xu G., Anal. Chim. Acta, 2022, 1221, 340116 |
| 18 | Lamanna J., Scott E. Y., Edwards H. S., Chamberlain M. D., Dryden M. D. M., Peng J., Mair B., Lee A., Chan C., Sklavounos A. A., Heffernan A., Abbas F., Lam C., Olson M. E., Moffat J., Wheeler A. R., Nat. Commun., 2020, 11(1), 5632 |
| 19 | Misra B. B., Methods Mol. Biol., 2020, 2064, 191—217 |
| 20 | Cao Y. Q., Zhang L., Zhang J., Guo Y. L., Anal. Chem., 2020, 92(12), 8378—8385 |
| 21 | Duncan K. D., Fyrestam J., Lanekoff I., Analyst, 2019, 144(3), 782—793 |
| 22 | Hu K., Nguyen T. D. K., Rabasco S., Oomen P. E., Ewing A. G., Anal. Chem., 2020, 93(1), 41—71 |
| 23 | Lageveen⁃Kammeijer G. S. M., de Haan N., Mohaupt P., Wagt S., Filius M., Nouta J., Falck D., Wuhrer M., Nat. Commun., 2019, 10(1), 2137 |
| 24 | Oomen P. E., Aref M. A., Kaya I., Phan N. T. N., Ewing A. G., Anal. Chem., 2018, 91(1), 588—621 |
| 25 | Yin L., Zhang Z., Liu Y., Gao Y., Gu J., Analyst, 2019, 144(3), 824—845 |
| 26 | Qin S., Zhang Y., Shi M., Miao D., Lu J., Wen L., Bai Y., Nat. Commun., 2024, 15(1), 4387 |
| 27 | Shao Y., Zhou Y., Liu Y., Zhang W., Zhu G., Zhao Y., Zhang Q., Yao H., Zhao H., Guo G., Zhang S., Zhang X., Wang X., Chem. Sci., 2022, 13(27), 8065—8073 |
| 28 | Taylor M. J., Mattson S., Liyu A., Stopka S. A., Ibrahim Y. M., Vertes A., Anderton C. R., Metabolites, 2021, 11(4), 200 |
| 29 | Chen A., Yan M., Feng J., Bi L., Chen L., Hu S., Hong H., Shi L., Li G., Jin B., Zhang X., Wen L., IEEE T Bio⁃Med Eng., 2022, 69(1), 325—333 |
| 30 | Marques C., Friedrich F., Liu L., Castoldi F., Pietrocola F., Lanekoff I., J. Am. Soc. Mass Spectrom., 2023, 34(11), 2518—2524 |
| 31 | Zhang W., Xu F., Yao J., Mao C., Zhu M., Qian M., Hu J., Zhong H., Zhou J., Shi X., Chen Y., Nat. Commun., 2023, 14(1), 2485 |
| 32 | Cao J., Yao Q. J., Wu J., Chen X., Huang L., Liu W., Qian K., Wan J. J., Zhou B. O., Cell Metab., 2024, 36(1), 209—221 |
| 33 | Fuchs S., Sawas N., Staedler N., Schubert D. A., D'Andrea A., Zeiser R., Piali L., Hruz P., Frei A. P., Eur. J. Immunol., 2019, 49(3), 462—475 |
| 34 | Palii C. G., Cheng Q., Gillespie M. A., Shannon P., Mazurczyk M., Napolitani G., Price N. D., Ranish J. A., Morrissey E., Higgs D. R., Brand M., Cell Stem Cell, 2019, 24(5), 812—820 |
| 35 | Wang B., Wang Y., Chen Y., Gao M., Ren J., Guo Y., Situ C., Qi Y., Zhu H., Li Y., Guo X., Brief Bioinform., 2022, 23(4), bbac214 |
| 36 | Zhang Y., Mol. Cell. Proteomics, 2023, 22(7), 100583 |
| 37 | Petelski A. A., Slavov N., Specht H., Jove⁃J. Vis. Exp., 2022,(190), e63802 |
| 38 | Shen B., Pade L. R., Choi S. B., Muñoz⁃Llancao P., Manzini M. C., Nemes P., Front. Chem., 2022, 10, 863979 |
| 39 | Slavov N., Development, 2023, 150(13), dev201492 |
| 40 | Subrahmanyam P. B., Maecker H. T., Methods Mol. Biol., 2021, 2285, 49—63 |
| 41 | Naval P., Paneda L. P., Athanasopoulou M., Teppo J. S., Zubarev R. A., Vegvari A., Methods Mol. Biol., 2024, 2817, 133—143 |
| 42 | Huffman R. G., Leduc A., Wichmann C., di Gioia M., Borriello F., Specht H., Derks J., Khan S., Khoury L., Emmott E., Petelski A. A., Perlman D. H., Cox J., Zanoni I., Slavov N., Nat. Methods, 2023, 20(5), 714—722 |
| 43 | Yuan Z., Zhou Q., Cai L., Pan L., Sun W., Qumu S., Yu S., Feng J., Zhao H., Zheng Y., Shi M., Li S., Chen Y., Zhang X., Zhang M. Q., Nat. Methods, 2021, 18(10), 1223—1232 |
| 44 | Jiang Y. R., Zhu L., Cao L. R., Wu Q., Chen J. B., Wang Y., Wu J., Zhang T. Y., Wang Z. L., Guan Z. Y., Xu Q. Q., Fan Q. X., Shi S. W., Wang H. F., Pan J. Z., Fu X. D., Wang Y., Fang Q., Cell Rep., 2023, 42(11), 113455 |
| 45 | Mennillo E., Kim Y. J., Lee G., Rusu I., Patel R. K., Dorman L. C., Flynn E., Li S., Bain J. L., Andersen C., Rao A., Tamaki S., Tsui J., Shen A., Lotstein M. L., Rahim M., Naser M., Bernard⁃Vazquez F., Eckalbar W., Cho S. J., Beck K., El⁃Nachef N., Lewin S., Selvig D. R., Terdiman J. P., Mahadevan U., Oh D. Y., Fragiadakis G. K., Pisco A., Combes A. J., Kattah M. G., Nat. Commun., 2024, 15(1), 1493 |
| 46 | Wu J., Xu Q. Q., Jiang Y. R., Chen J. B., Ying W. X., Fan Q. X., Wang H. F., Wang Y., Shi S. W., Pan J. Z., Fang Q., Anal. Chem., 2024, 96(14), 5499—5508 |
| 47 | Zhao P., Feng Y., Wu J., Zhu J., Yang J., Ma X., Ouyang Z., Zhang X., Zhang W., Wang W., Anal. Chem., 2023, 95(18), 7212—7219 |
| 48 | Wagner J., Rapsomaniki M. A., Chevrier S., Anzeneder T., Langwieder C., Dykgers A., Rees M., Ramaswamy A., Muenst S., Soysal S. D., Cell, 2019, 177(5), 1330—1345 |
| 49 | Friebel E., Kapolou K., Unger S., Núñez N. G., Utz S., Rushing E. J., Regli L., Weller M., Greter M., Tugues S., Cell, 2020, 181(7), 1626—1642 |
| 50 | Guo Y., Cai L., Liu X., Ma L., Zhang H., Wang B., Qi Y., Liu J., Diao F., Sha J., Mol. Cell. Proteomics, 2022, 21(8), 100267 |
| 51 | Schoof E. M., Furtwängler B., Üresin N., Rapin N., Savickas S., Gentil C., Lechman E., Keller U. A. D., Dick J. E., Porse B. T., Nat. Commun., 2021, 12(1), 3341 |
| 52 | Brunner A. D., Thielert M., Vasilopoulou C., Ammar C., Coscia F., Mund A., Hoerning O. B., Bache N., Apalategui A., Lubeck M., Mol. Syst. Biol., 2022, 18(3), e10798 |
| 53 | Tsai A. G., Glass D. R., Juntilla M., Hartmann F. J., Oak J. S., Fernandez⁃Pol S., Ohgami R. S., Bendall S. C., Nat. Med., 2020, 26(3), 408—417 |
| 54 | Zhang L., Du F., Jin Q., Sun L., Wang B., Tan Z., Meng X., Huang B., Zhan Y., Su W., Adv. Sci., 2023, 10(35), 2300123 |
| 55 | Georg P., Astaburuaga⁃García R., Bonaguro L., Brumhard S., Michalick L., Lippert L. J., Kostevc T., Gäbel C., Schneider M., Streitz M., Cell, 2022, 185(3), 493—512 |
| 56 | Mogilenko D. A., Shpynov O., Andhey P. S., Arthur L., Swain A., Esaulova E., Brioschi S., Shchukina I., Kerndl M., Bambouskova M., Immunity, 2021, 54(1), 99—115 |
| [1] | 侯泽金, 李荣其, 李健, 冯怡宁, 靳茜茜, 孙俊红, 曹洁. 基于GC-MS和机器学习的深静脉血栓形成预测[J]. 高等学校化学学报, 2024, 45(9): 20240199. |
| [2] | 刘康, 潘荣容, 江德臣. 基于纳米电极的单细胞内生物分子电化学分析[J]. 高等学校化学学报, 2024, 45(5): 20240027. |
| [3] | 董沛滢, 刘彤, 秦伟捷. RNA-蛋白质复合物规模化富集与鉴定新方法[J]. 高等学校化学学报, 2024, 45(11): 20240091. |
| [4] | 张磊, 申华莉. 标志物研究中常用蛋白组学质谱方法的定量准确性评估[J]. 高等学校化学学报, 2024, 45(11): 20240311. |
| [5] | 靳莹, 张俊杰, 张毅欣, 袁悦, 韩珍珍. 外泌体分离和蛋白质组学分析的研究进展[J]. 高等学校化学学报, 2024, 45(11): 20240305. |
| [6] | 胡宇虹, 俞相明, 宋丽丽, 邢清和, 周峰. DEEP SEQ方法检测新生儿毛干蛋白质组动态变化[J]. 高等学校化学学报, 2024, 45(11): 20240326. |
| [7] | 范智瑞, 方群, 杨奕. 基于质谱的单细胞蛋白质组学分析[J]. 高等学校化学学报, 2024, 45(11): 20240294. |
| [8] | 曹宜青, 侯静欣, 刘建业, 李嫣. 体液外泌体代谢组学研究进展和挑战[J]. 高等学校化学学报, 2024, 45(11): 20240324. |
| [9] | 许霞, 秦伟达, 李若萌, 王倩倩, 刘宁, 李功玉. 深度覆盖蛋白质组学质谱分析: 细胞蛋白提取方法的评估[J]. 高等学校化学学报, 2024, 45(11): 20240344. |
| [10] | 黄玉滢, 于成鲲, 刘斯奇, 任艳. 利用无标记单细胞蛋白质组学方法构建小鼠外周血单个核细胞的细胞图谱[J]. 高等学校化学学报, 2024, 45(11): 20240355. |
| [11] | 沈枫林, 冯兆莹, 方静, 张磊, 刘晓慧, 周新文. 基于质谱的单细胞分辨的空间蛋白质组学新技术研究[J]. 高等学校化学学报, 2024, 45(11): 20240299. |
| [12] | 赵雪琪, 赵越, 薛静, 白敏, 陈锋, 孙颖, 宋大千, 赵永席. 单细胞核酸编码扩增成像分析[J]. 高等学校化学学报, 2022, 43(12): 20220572. |
| [13] | 赵欢欢, 李存玉, 金红. 基于TMT10-plex等量标记结合SMOAC富集磷酸肽的定量磷酸化蛋白质组性能评价[J]. 高等学校化学学报, 2021, 42(12): 3624. |
| [14] | 张楠茜, 吕经纬, 金平, 李晶峰, 边学峰, 张辉, 孙佳明. N-苄基十六碳酰胺促小鼠Leydig细胞增殖和分泌睾酮的 1H NMR代谢组学研究[J]. 高等学校化学学报, 2019, 40(9): 1832. |
| [15] | 李雯雯, 朱爱如, 龙怡静, 王春燕, 韩源平, 段忆翔. 高脂与维生素D缺乏饮食诱导的2型糖尿病小鼠血清和肝脏代谢组学研究[J]. 高等学校化学学报, 2018, 39(11): 2395. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||