

Chem. J. Chinese Universities ›› 2020, Vol. 41 ›› Issue (12): 2587.doi: 10.7503/cjcu20200592
Previous Articles Next Articles
LI Xianming1, ZHENG Ting2, GAO Lu2, LI Feng1,2( ), HOU Xiandeng1,2, WU Peng1,2(
), HOU Xiandeng1,2, WU Peng1,2( )
)
Received:2020-08-23
Online:2020-12-10
Published:2020-12-09
Contact:
LI Feng
E-mail:windtalker_1205@scu.edu.cn;wupeng@scu.edu.cn
Supported by:CLC Number:
TrendMD:
LI Xianming, ZHENG Ting, GAO Lu, LI Feng, HOU Xiandeng, WU Peng. Recombinase Polymerase Amplification: from Principle to Performance[J]. Chem. J. Chinese Universities, 2020, 41(12): 2587.
| Category | Strategy I | Strategy II | Function description |
|---|---|---|---|
| Recombinase | uvsX | RecA | DNA?dependent ATPase, the key protein for DNA strand exchange |
ssDNA binding proteins | gp32 | SSB | Melting the secondary structure of primers to decrease undesirable interactions |
| Accessory proteins | uvsY | RecF, RecO | Assisting the loading of recombinase on primer |
| Crowding agents | PEGs, dectran, ficoll | Accelerating the RP process and primer extending | |
| Polymerase | T4 polymerase, Bsu DNA polymerase I, etc. | Extending the primer and displacement of the complementary strand of templates | |
| ATP regeneration | Phosphocreatine and creatine | Transforming ADP into ATP | |
| Reverse transcriptase | Mu?MLV reverse transcriptase | Converting RNA templates to ssDNA | |
Table 1 Functional substances involved in recombinase polymerase amplification
| Category | Strategy I | Strategy II | Function description |
|---|---|---|---|
| Recombinase | uvsX | RecA | DNA?dependent ATPase, the key protein for DNA strand exchange |
ssDNA binding proteins | gp32 | SSB | Melting the secondary structure of primers to decrease undesirable interactions |
| Accessory proteins | uvsY | RecF, RecO | Assisting the loading of recombinase on primer |
| Crowding agents | PEGs, dectran, ficoll | Accelerating the RP process and primer extending | |
| Polymerase | T4 polymerase, Bsu DNA polymerase I, etc. | Extending the primer and displacement of the complementary strand of templates | |
| ATP regeneration | Phosphocreatine and creatine | Transforming ADP into ATP | |
| Reverse transcriptase | Mu?MLV reverse transcriptase | Converting RNA templates to ssDNA | |
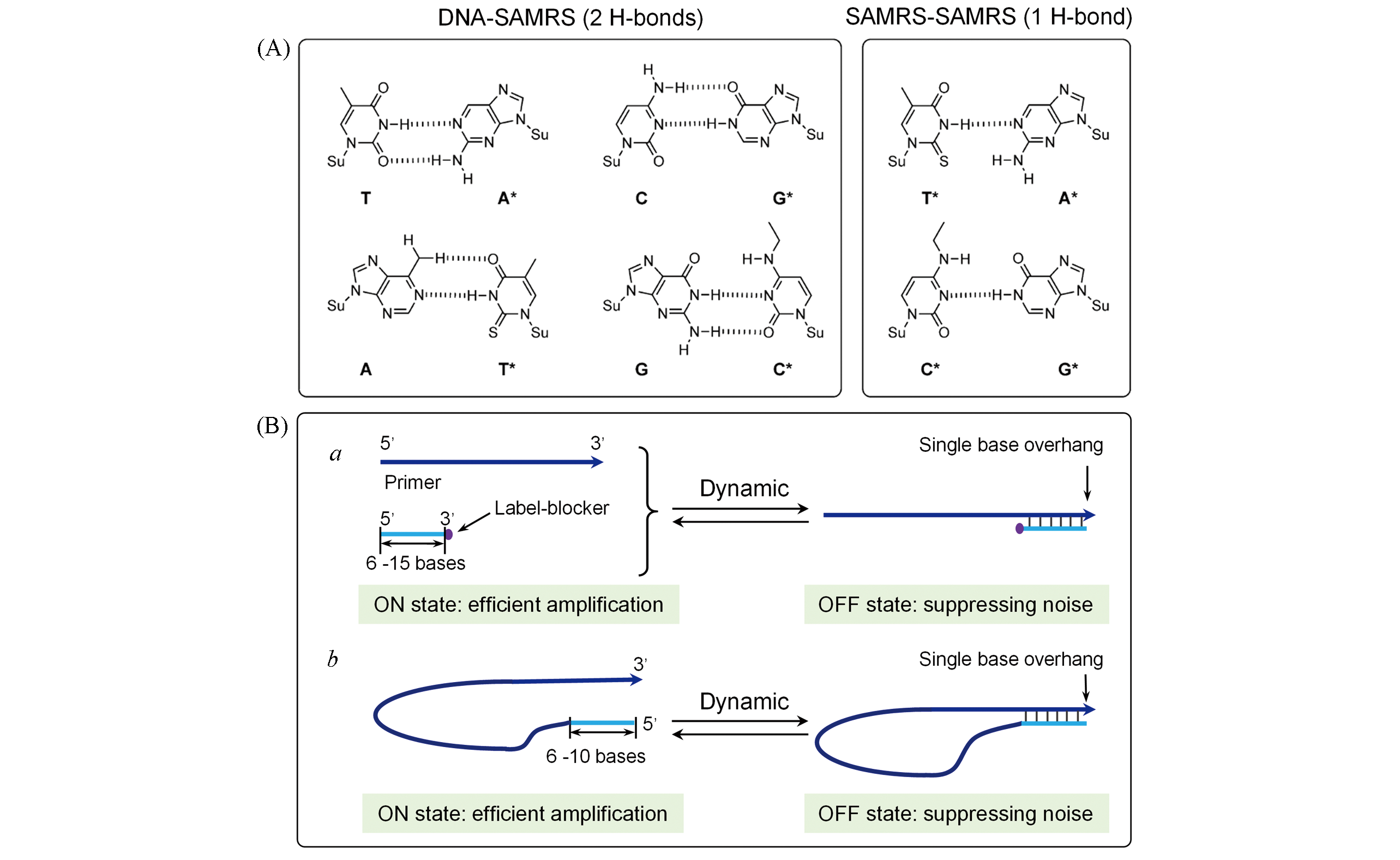
Fig.3 Primers for reducing nose products(A) Chemical formulas of the self?avoiding molecular?recognition system[33], Copyright 2014, Wiley?VCH;(B) competition designs.
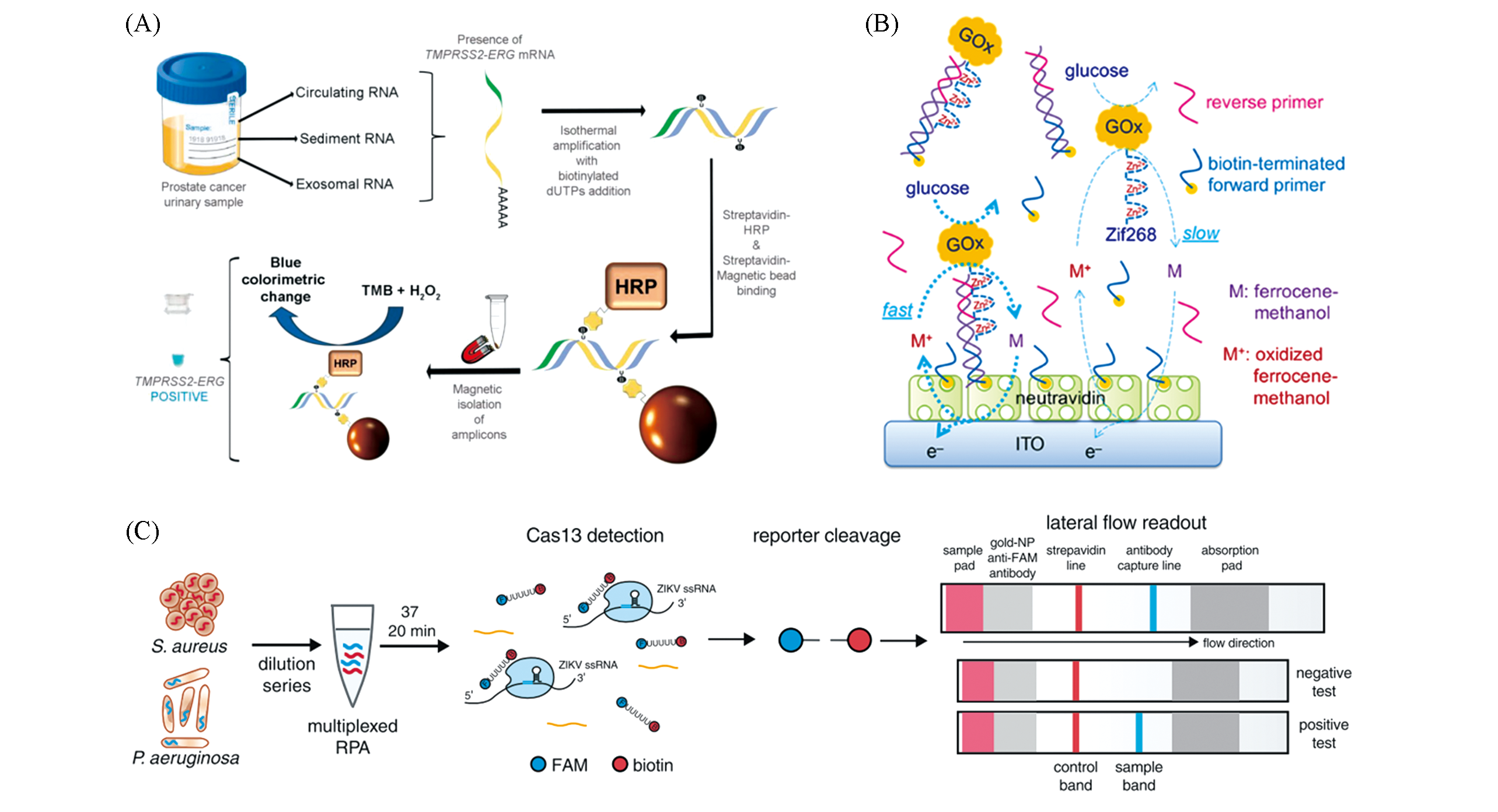
Fig.5 New amplicon testing methods(A) Colorimetric assay[45], Copyright 2018, Royal Society of Chemistry; (B) electrochemical method[46], Copyright 2018, Royal Society of Chemistry; (C) CTISPR/Cas based lateral flow readout[48], Copyright 2018 American Association for the Advancement of Science.
| Assaya | Primer number | Temperature/℃ | Reaction/min | Detection formatsb | Target | Reagent format | Multiplex | Ref. |
|---|---|---|---|---|---|---|---|---|
| NASBA | 2 | 41 | 105 | RTF, NALF | RNA(DNA) | Liquid | Y | [ |
| TMA | 2 | 60 | 140 | RTF | RNA(DNA) | Liquid | Y | [ |
| SMART | 2 | 41 | 140 | RTF | RNA, DNA | Liquid | N/A | [ |
| LAMP | 4 | 60—65 | 60—90 | RTF, NALF, RTT, TE | RNA, DNA | Liquid | N/A | [ |
| CPA | 4 | 65 | 65 | RTF, NALF | DNA | Liquid | N/A | [ |
| RCA | 1, 2 | 65 | 60 | RTF | RNA, DNA | Liquid | N/A | [ |
| RAM | 1 | 63 | 120—180 | RTF | RNA, DNA | Liquid | N/A | [ |
| SDA | 2 | 37 | 120 | RTF, NALF | RNA, DNA | Liquid | Y | [ |
| NEAR | 4 | 55 | 10 | RTF, NALF | RNA, DNA | Liquid | Y | [ |
| NEMA | 4 | 65 | 30 | NALF | DNA | Liquid | N/A | [ |
| ICA | 2 | 60 | 60 | RTF | DNA | Liquid | N/A | [ |
| EXPAR | 1 | 55 | 10—20 | RTF, NALF | DNA | Liquid | Y | [ |
| HDA | 2 | 65 | 75—90 | RTF, NALF | RNA, DNA | Liquid | Y | [ |
| RPA | 2 | 30—42 | 20 | RTF, NALF | RNA, DNA | Liquid, lyophilized | Y | [ |
Table 2 Comparison of current isothermal amplification methods
| Assaya | Primer number | Temperature/℃ | Reaction/min | Detection formatsb | Target | Reagent format | Multiplex | Ref. |
|---|---|---|---|---|---|---|---|---|
| NASBA | 2 | 41 | 105 | RTF, NALF | RNA(DNA) | Liquid | Y | [ |
| TMA | 2 | 60 | 140 | RTF | RNA(DNA) | Liquid | Y | [ |
| SMART | 2 | 41 | 140 | RTF | RNA, DNA | Liquid | N/A | [ |
| LAMP | 4 | 60—65 | 60—90 | RTF, NALF, RTT, TE | RNA, DNA | Liquid | N/A | [ |
| CPA | 4 | 65 | 65 | RTF, NALF | DNA | Liquid | N/A | [ |
| RCA | 1, 2 | 65 | 60 | RTF | RNA, DNA | Liquid | N/A | [ |
| RAM | 1 | 63 | 120—180 | RTF | RNA, DNA | Liquid | N/A | [ |
| SDA | 2 | 37 | 120 | RTF, NALF | RNA, DNA | Liquid | Y | [ |
| NEAR | 4 | 55 | 10 | RTF, NALF | RNA, DNA | Liquid | Y | [ |
| NEMA | 4 | 65 | 30 | NALF | DNA | Liquid | N/A | [ |
| ICA | 2 | 60 | 60 | RTF | DNA | Liquid | N/A | [ |
| EXPAR | 1 | 55 | 10—20 | RTF, NALF | DNA | Liquid | Y | [ |
| HDA | 2 | 65 | 75—90 | RTF, NALF | RNA, DNA | Liquid | Y | [ |
| RPA | 2 | 30—42 | 20 | RTF, NALF | RNA, DNA | Liquid, lyophilized | Y | [ |
| 1 | Wang R. F., Cao W. W., Cerniglia C. E., J. Appl. Microbiol., 1997, 83(6), 727—736 |
| 2 | Bustin Stephen A., Mueller R., Clin. Sci., 2005, 109(4), 365—379 |
| 3 | Morling N., Biochem. Soc. Trans., 2009, 37(2), 438—440 |
| 4 | Clerc O., Greub G., Clin. Microbiol. Infect., 2010, 16(8), 1054—1061 |
| 5 | Dong T. Y., Mansour H., Hu H., Wang G. A., Watson C. J. F., Yousef M., Matamoros G., Sanchez A. L., MacNeil A. J., Wu P., Li F., Anal. Chem., 2020, 92(9), 6456—6461 |
| 6 | Daher R. K., Stewart G., Boissinot M., Bergeron M. G., Clin. Chem., 2016, 947—958 |
| 7 | Lobato I. M., O’Sullivan C. K., Trends Anal. Chem., 2017, 19—35 |
| 8 | Li J., Macdonald J., von Stetten F., Analyst, 2019, 144(1), 31—67 |
| 9 | Stringer O. W., Andrews J. M., Greetham H. L., Forrest M. S., Nat. Methods, 2018, 15(5), I—III |
| 10 | Piepenburg O., Williams C. H., Stemple D. L., Armes N. A., PLoS Biol., 2006, 4(7), e204 |
| 11 | Zaghloul H., El⁃shahat M., World J. Hepatol., 2014, 6(12), 916—922 |
| 12 | James A., Macdonald J., Expert Rev. Mol. Diagn., 2015, 15(11), 1475—1489 |
| 13 | Moore M. D., Jaykus L. A., Future Virol., 2017, 12(8), 421—429 |
| 14 | Babu B., Ochoa⁃Corona F. M., Paret M. L., Anal. Biochem., 2018, 546, 72—77 |
| 15 | Piepenburg O., Williams Colin H., Armes Niall A., Stemple Derek L., Recombinase Polymerase Amplification, US 20190360030, 2019⁃11⁃28 |
| 16 | Kowalczykowski S. C., Krupp R. A., J. Mol. Biol., 1987, 193(1), 97—113 |
| 17 | Bar⁃Ziv R., Libchaber A., Proc. Natl. Acad. Sci. USA, 2001, 98(16), 9068—9073 |
| 18 | Roy R., Kozlov A. G., Lohman T. M., Ha T., Nature, 2009, 461(7267), 1092—1097 |
| 19 | Liu J., Morrical S. W., Virol J., 2010, 7(1), 357 |
| 20 | Mok E., Wee E., Wang Y., Trau M., Sci. Rep., 2016, 6(1), 37837 |
| 21 | Bianco P. R., Tracy R. B., Kowalczykowski S. C., Front. Biosci., 1998, 3, 570—603 |
| 22 | Bell J. C., Kowalczykowski S. C., Trends Biochem. Sci., 2016, 41(6), 491—507 |
| 23 | Kowalezykowski S. C., Annu. Rev. Biophys. Biophys. Chem., 1991, 20(1), 539—575 |
| 24 | Rosselli W., Stasiak A., EMBO J., 1991, 10(13), 4391—4396 |
| 25 | Danilowicz C., Hermans L., Coljee V., Prévost C., Prentiss M., Nucleic Acids Res., 2017, 45(14), 8448—8462 |
| 26 | Lee J. Y., Qi Z., Greene E. C., J. Biol. Chem., 2016, 291(42), 22218—22230 |
| 27 | Bittl J. A., Ingwall J. S., J. Biol. Chem., 1985, 260(6), 3512—3517 |
| 28 | Zhao B., Zhang D., Li C., Yuan Z., Yu F., Zhong S., Jiang G., Yang Y. G., Le X. C., Weinfeld M., Zhu P., Wang H., Cell Discov., 2017, 3(1), 16053 |
| 29 | Kim S. H., Ragunathan K., Park J., Joo C., Kim D., Ha T., J. Am. Chem. Soc., 2014, 136(42), 14796—14800 |
| 30 | Morrical S. W., Alberts B. M., J. Biol. Chem., 1990, 265(25), 15096—15103 |
| 31 | Cox M. M., Crit. Rev. Biochem. Mol. Biol., 2007, 42(1), 41—63 |
| 32 | Cox M. M., The Bacterial RecA Protein: Structure, Function, and Regulation, Springer Berlin Heidelberg, Berlin, 2007, 53—94 |
| 33 | Sharma N., Hoshika S., Hutter D., Bradley K. M., Benner S. A., ChemBioChem, 2014, 15(15), 2268—2274 |
| 34 | Higgins M., Ravenhall M., Ward D., Phelan J., Ibrahim A., Forrest M. S., Clark T. G., Campino S., Bioinformatics, 2018, 35(4), 682—684 |
| 35 | del Río J. S., Adly N. Y., Acero⁃Sánchez J. L., Henry O. Y., O’Sullivan C. K., Biosens. Bioelectron., 2014, 54, 674—678 |
| 36 | Crannell Z. A., Cabada M. M., Castellanos⁃Gonzalez A., Irani A., White A. C., Richards⁃Kortum R., Am. J. Trop. Med. Hyg., 2015, 92(3), 583—587 |
| 37 | Kunze A., Dilcher M., Abd El Wahed A., Hufert F., Niessner R., Seidel M., Anal. Chem., 2015, 88(1), 898—905 |
| 38 | Ng B. Y., Wee E. J., West N. P., Trau M., Sci. Rep., 2015, 5, 15027 |
| 39 | Kim J., Biondi M. J., Feld J. J., Chan W. C., ACS Nano, 2016, 10(4), 4742—4753 |
| 40 | Liu H. B., Du X. J., Zang Y. X. Li P., Wang S., J. Agric. Food Chem., 2017, 65(47), 10290—10299 |
| 41 | Chertow D. S., Science, 2018, 360(6387), 381—382 |
| 42 | Shin Y., Perera A. P., Kim K. W., Park M. K., Lab Chip, 2013, 13(11), 2106—2114 |
| 43 | Lau H. Y., Wang Y., Wee E. J., Botella J. R., Trau M., Anal. Chem., 2016, 88(16), 8074—8081 |
| 44 | Ng B. Y., Wee E. J., West N. P., Trau M., ACS Sens., 2015, 1(2), 173—178 |
| 45 | Islam M. N., Moriam S., Umer M., Phan H. P., Salomon C., Kline R., Nguyen N. T., Shiddiky M. J., Analyst, 2018, 3021—3028 |
| 46 | Fang C. S., Kim K. S., Ha D. T., Kim M. S., Yang H., Anal. Chem., 2018, 90(7), 4776—4782 |
| 47 | Gootenberg J. S., Abudayyeh O. O., Lee J. W., Essletzbichler P., Dy A. J., Joung J., Verdine V., Donghia N., Daringer N. M., Freije C. A., Science, 2017, 356(6336), 438—442 |
| 48 | Gootenberg J. S., Abudayyeh O. O., Kellner M. J., Joung J., Collins J. J., Zhang F., Science, 2018, 360(6387), 439—444 |
| 49 | Chen J. S., Ma E., Harrington L. B., Da Costa M., Tian X., Palefsky J. M., Doudna J. A., Science, 2018, 360(6387), 436—439 |
| 50 | Myhrvold C., Freije C. A., Gootenberg J. S., Abudayyeh O. O., Metsky H. C., Durbin A. F., Kellner M. J., Tan A. L., Paul L. M., Parham L. A., Science, 2018, 360(6387), 444—448 |
| 51 | Kunze A., Dilcher M., Abd El Wahed A., Hufert F., Niessner R., Seidel M., Anal. Chem., 2015, 88(1), 898—905 |
| 52 | Kober C., Niessner R., Seidel M.,. Biosens. Bioelectron., 2018, 100, 49—55 |
| 53 | Elsäßer D., Ho J., Niessner R., Tiehm A., Seidel M., Anal. Biochem., 2018, 546, 58—64 |
| 54 | Lau H. Y., Wang Y., Wee E. J., Botella J. R., Trau M., Anal. Chem., 2016, 88(16), 8074—8081 |
| 55 | Koo K. M. Wee E. J., Mainwaring P. N., Wang Y., Trau M., Small, 2016, 12(45), 6233—6242 |
| 56 | Gracias K. S., Mckillip J. L., J. Rapid Methods Autom. Microbiol., 2007, 15(3), 295—309 |
| 57 | Hofmann W. P., Dries V., Herrmann E., Gärtner B., Zeuzem S., Sarrazin C., J. Clin. Virol., 2005, 32(4), 289—293 |
| 58 | Wharam S. D., Hall M. J., Wilson W. H., Virol. J., 2007, 4(1), 52 |
| 59 | Curtis K. A., Rudolph D. L., Owen S. M., J. Med. Virol., 2009, 81(6), 966—972 |
| 60 | Mori Y., Notomi T., J. Infect. Chemother., 2009, 15(2), 62—69 |
| 61 | Fang R., Li X., Hu L., You Q., Li J., Wu J., Xu P., Zhong H., Luo Y., Mei J., Gao Q., J. Clin. Microbiol., 2009, 47(3), 845—847 |
| 62 | Johne R., Müller H., Rector A., van Ranst M., Stevens H., Trends Microbiol., 2009, 17(5), 205—211 |
| 63 | Yao B., Li J., Huang H., Sun C., Wang Z., Fan Y., Chang Q., Li S., Xi J., RNA, 2009, 15(9), 1787—1794 |
| 64 | Hellyer T. J., Nadeau J. G., Expert Rev. Mol. Diagn., 2004, 4(2), 251—261 |
| 65 | McHugh T. D., Pope C. F., Ling C. L., Patel S., Billington O. J., Gosling R. D., Lipman M. C., Gillespie S. H., J. Med. Microbiol., 2004, 53(12), 1215—1219 |
| 66 | Maples B. K., Holmberg R. C., Miller A. P.,Provins J., Rot R., Mandell J., Nicking and Extension Amplification Reaction for the Exponential Amplification of Nucleic Acids, US 9562264, 2017⁃07⁃02 |
| 67 | You Q. M., Hu L., Wan J., Zhong H. Y., Method for Amplifying Target Nucleic Acid Sequence by Nickase, and Kit for Amplifying Target Nucleic Acid Sequence and Its Use, CP 1811447A, 2006⁃08⁃02 |
| 68 | Jung C., Chung J. W., Kim U. O., Kim M. H., Park H. G., Anal. Chem., 2010, 82(14), 5937—5943 |
| 69 | van Ness J., van Ness L. K., Galas D. J., Proc. Natl. Acad. Sci. USA, 2003, 100(8), 4504—4509 |
| 70 | Tan E., Erwin B., Dames S., Voelkerding K., Niemz A., Clin. Chem., 2007, 53(11), 2017—2020 |
| 71 | Jeong Y. J., Park K., Kim D. E., Cell. Mol. Life Sci., 2009, 66(20), 3325 |
| 72 | Vincent M., Xu Y., Kong H., EMBO Rep., 2004, 5(8), 795—800 |
| 73 | Lutz S., Weber P., Focke M., Faltin B., Hoffmann J., Müller C., Mark D., Roth G., Munday P., Armes N., Piepenburg O., Zengerle R., von Stetten F., Lab Chip, 2010, 10(7), 887—893 |
| 74 | Koo K. M., Wee E. J., Trau M., Theranostics, 2016, 6(9), 1415—1424 |
| 75 | Hu C., Kalsi S., Zeimpekis I., Sun K., Ashburn P., Turner C., Sutton J. M. Morgan H., Biosens. Bioelectron., 2017, 96, 281—287 |
| 76 | Koo K. M., Wee E. J., Trau M., Biosens. Bioelectron., 2017, 89, 715—720 |
| 77 | Ng B. Y., Wee E. J., Woods K., Anderson W., Antaw F., Tsang H. Z., West N. P., Trau M., Anal. Chem., 2017, 89(17), 9017—9022 |
| 78 | James A., Macdonald J., Expert Rev. Mol. Diagn., 2015, 15(11), 1475—1489 |
| 79 | Kissenkötter J., Böhlken⁃Fascher S., Forrest M. S., Piepenburg O., Czerny C. P., Abd El Wahed A., Food Chem., 2020, 322, 126759 |
| 80 | Kalsi S., Valiadi M., Tsaloglou M. N., Parry⁃Jones L., Jacobs A., Watson R., Turner C., Amos R., Hadwen B., Buse J., Lab Chip, 2015, 15(14), 3065—3075 |
| 81 | Yeh E. C., Fu C. C., Hu L., Thakur R., Feng J., Lee L. P., Sci. Adv., 2017, 3(3), e1501645 |
| 82 | Li J., Ma B., Fang J., Zhi A., Chen E., Xu Y., Yu X., Sun C., Zhang M., Foods, 2020, 9(1), 27 |
| 83 | Lutz S., Weber P., Focke M., Faltin B., Hoffmann J., Müller C., Mark D., Roth G., Munday P., Armes N., Piepenburg O., Zengerle R., von Stetten F., Lab Chip, 2010, 10(7), 887—893 |
| 84 | Foudeh A. M., Fatanat Didar T., Veres T., Tabrizian M., Lab Chip, 2012, 12(18), 3249—3266 |
| 85 | Kim T. H., Park J., Kim C. J., Cho Y. K., Anal. Chem., 2014, 86(8), 3841—3848 |
| 86 | Chen J., Xu Y., Yan H., Zhu Y., Wang L., Zhang Y., Lu Y., Xing W., Lab Chip, 2018, 18(16), 2441—2452 |
| 87 | Rohrman B. A., Richards⁃Kortum R. R., Lab Chip, 2012, 12(17), 3082—3088 |
| 88 | Ahn H., Batule B. S., Seok Y., Kim M. G., Anal. Chem., 2018, 90(17), 10211—10216 |
| 89 | Ming K., Kim J., Biondi M. J., Syed A., Chen K., Lam A., Ostrowski M., Rebbapragada A., Feld J. J., Chan W. C., ACS Nano, 2015, 9(3), 3060—3074 |
| 90 | Amaratunga M., Benight A. S., Biochem. Biophys. Res. Commun., 1988, 157(1), 127—133 |
| 91 | Kim S. H., Joo C., Ha T., Kim D., Nucleic Acids Res., 2013, 41(16), 7738—7744 |
| 92 | Wang Y., Zhou R., Liu W., Liu C., Wu P., Chin. Chem. Lett., 2020, doi: 10.1016/j.cclet.2020.01.023 |
| 93 | Butler B. C., Hanchett R. H., Rafailov H., MacDonald G., Biophys. J., 2002, 82(4), 2198—2210 |
| 94 | Lenhart J. S., Brandes E. R., Schroeder J. W., Sorenson R. J., Showalter H. D., Simmons L. A., J. Bacteriol., 2014, 196(15), 2851—2860 |
| 95 | Drain P. K., Hyle E. P., Noubary F., Freedberg K. A., Wilson D., Bishai W. R., Rodriguez W., Bassett I. V., Lancet Infect. Dis., 2014, 14(3), 239—249 |
| [1] | LIU Suyu, DING Fei, LI Qian, FAN Chunhai, FENG Jing. Azobenzene-integrated DNA Nanomachine [J]. Chem. J. Chinese Universities, 2022, 43(8): 20220122. |
| [2] | WU Yushuai, SHANG Yingxu, JIANG Qiao, DING Baoquan. Research Progress of Controllable Self-assembled DNA Origami Structure as Drug Carrier [J]. Chem. J. Chinese Universities, 2022, 43(8): 20220179. |
| [3] | WANG Junyang, LIU Zheng, ZHANG Qian, SUN Chunyan, LI Hongxia. Application of DNA Silver Nanoclusters in the Fluorescence Biosensors based on Functional Nucleic Acids [J]. Chem. J. Chinese Universities, 2022, 43(6): 20220010. |
| [4] | WEI Minmin, YUAN Ze, LU Min, MA Hui, XIE Xiaoji, HUANG Ling. Recent Advances in Lanthanide Doped Upconversion Nanoparticle-Metal Organic Framework Composites [J]. Chem. J. Chinese Universities, 2021, 42(8): 2313. |
| [5] | WU Yangyi, CHEN Jianping, Ai Yijing, WANG Qingxiang, GAO Fei, GAO Feng. Synthesis of 2-(2-Hydroxy-3-methoxyphenyl)-C60 and Its Application for Sensing of Cauliflower Mosaic Virus 35S Promotor [J]. Chem. J. Chinese Universities, 2021, 42(6): 1754. |
| [6] | GE Haoying, DU Jianjun, LONG Saran, SUN Wen, FAN Jiangli, PENG Xiaojun. Surface Functionalized Gold Nanomaterials in Tumor Diagnosis and Treatment [J]. Chem. J. Chinese Universities, 2021, 42(4): 1202. |
| [7] | CHEN Hongda, ZHANG Hua, WANG Zhenxin. Development of Small Animals in vivo Fluorescence-photothermal Dual Mode Imaging System [J]. Chem. J. Chinese Universities, 2021, 42(3): 725. |
| [8] | YANG Xinjie, LAI Yanqiong, LI Qiuyang, ZHANG Yanli, WANG Hongbin, PANG Pengfei, YANG Wenrong. An Enzyme-free and Label-free Fluorescent Probe for Detection of Microcystin-LR Based on Circular DNA-Silver Nanoclusters [J]. Chem. J. Chinese Universities, 2021, 42(12): 3600. |
| [9] | HU Ling, YIN Yao, KE Guoliang, ZHANG Xiaobing. Regulation of Cell-cell Interactions Based on DNA Nanostructures [J]. Chem. J. Chinese Universities, 2021, 42(11): 3284. |
| [10] | MAO Yu, QU Hao, ZHENG Lei. Research Progress on RNA⁃cleaving DNAzyme for the Detection of Pathogenic Bacteria [J]. Chem. J. Chinese Universities, 2021, 42(11): 3445. |
| [11] | WANG Qing, HE Yuqiu, WANG Fuan. Advances of Multifunctional Deoxyribozyme in Biomedical Analysis [J]. Chem. J. Chinese Universities, 2021, 42(11): 3334. |
| [12] | XI Jing, CHEN Na, YANG Yanbing, YUAN Quan. Recent Progress in Controlled Synthesis of Persistent Luminescence Nanomaterials for Diagnosis Applications [J]. Chem. J. Chinese Universities, 2021, 42(11): 3247. |
| [13] | LIU Xuejiao, YANG Fan, LIU Shuang, ZHANG Chunjuan, LIU Qiaoling. Progress in Aptamer-targeted Membrane Protein Recognition and Functional Regulation [J]. Chem. J. Chinese Universities, 2021, 42(11): 3277. |
| [14] | LIU Yuan, DENG Jinqi, ZHAO Shuai, TIAN Fei, LI Yi, SUN Jiashu, LIU Chao. Lateral Flow Assay Based on Molecular Recognition for Diagnosis of Corona Virus Disease 2019 Infection [J]. Chem. J. Chinese Universities, 2021, 42(11): 3390. |
| [15] | PENG Huo, GAO Zehang, LIAO Chengyue, WANG Xiaodong, ZHOU Hongbo, ZHAO Jianlong. Robust Droplet Digital PCR Chip for Absolute Quantitative Detection of Nucleic Acid [J]. Chem. J. Chinese Universities, 2020, 41(8): 1760. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||