

Chem. J. Chinese Universities ›› 2024, Vol. 45 ›› Issue (11): 20240345.doi: 10.7503/cjcu20240345
• Article • Previous Articles Next Articles
JIANG Yan1, CHEN Yanlin2, SONG Gaoyu3, CHEN Yanyan4, BAI Jing1, ZHU Yingdi3( ), LI Juan3(
), LI Juan3( )
)
Received:2024-07-08
Online:2024-11-10
Published:2024-07-28
Contact:
ZHU Yingdi, LI Juan
E-mail:zhuyingdi@him.cas.cn;lijuan@him.cas.cn
Supported by:CLC Number:
TrendMD:
JIANG Yan, CHEN Yanlin, SONG Gaoyu, CHEN Yanyan, BAI Jing, ZHU Yingdi, LI Juan. Bacterial Protein Profiling[J]. Chem. J. Chinese Universities, 2024, 45(11): 20240345.
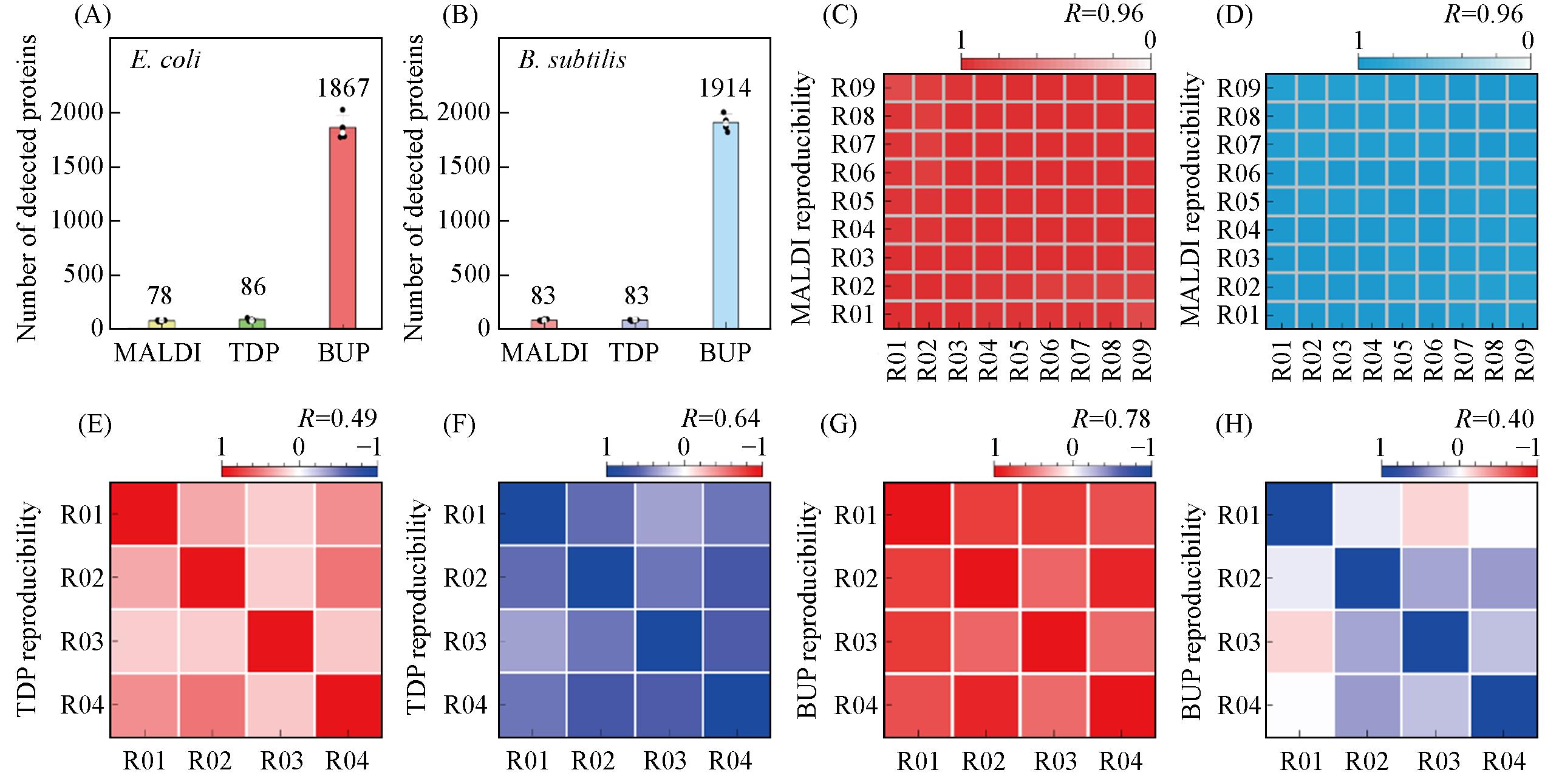
Fig.2 Investigation of protein coverage and detection reproducibility(A) Numbers of proteins or protein peaks detected from E. coli by MALDI fingerprinting(MALDI), top-down proteomics(TDP) and bottom-up proteomics(BUP); the average number was displayed for each approach; (B) numbers of proteins or protein peaks detected from B. subtilis by using the three protein profiling approaches; (C) detection reproducibility for nine repetition tests of E. coli by using MALDI approach; (D) detection reproducibility for nine repetition tests of B. subtilis by using MALDI approach; (E) detection reproducibility for four repetition tests of E. coli by using TDP approach; (F) detection reproducibility for four repetition tests of B. subtilis by using TDP approach; (G) detection reproducibility for four repetition tests of E. coli by using BUP approach; (H) detection reproducibility for four repetition tests of B. subtilis by using BUP approach.
| MALDI(m/z) | TDP precursormass | Protein entry name | Gene ID | Subcellularlocation | Isoelectricpoint | Instability index | Aliphaticindex | GRAVY | Detected by BUP |
|---|---|---|---|---|---|---|---|---|---|
| 6404 | 6406.60 | P0AG51_RL30 | rpmD | Cytoplasm | 10.96 | 38.72 | 102.59 | -0.137 | YES |
| 6849 | 6848.53 | P0AAZ7_YCAR | ycaR | Cytoplasm | 4.31 | 36.22 | 105.39 | -0.127 | YES |
| 7264 | 7268.98 | P0A7M6_RL29 | rpmC | Cytoplasm | 9.98 | 50.27 | 102.22 | -0.657 | YES |
| 7861 | 7865.92 | P0A7M9_RL31 | rpmE | Cytoplasm | 9.46 | 48.68 | 57.00 | -0.649 | NO |
| 8107 | 8113.28 | P69222_IF1 | infA | Cytoplasm | 9.23 | 36.00 | 91.83 | -0.354 | NO |
| 8354 | 8344.10 | P68206_YJBJ | yjbJ | Cytoplasm/Periplasm | 5.64 | 38.38 | 15.60 | -2.432 | NO |
| 8871 | 8869.83 | P0A7M2_RL28 | rpmB | Cytoplasm | 11.42 | 36.21 | 77.87 | -0.650 | YES |
| 9213 | 9219.99 | P0ACF4_DBHB | hupB | Cytoplasm/Periplasm | 9.69 | 16.40 | 92.44 | -0.042 | YES |
| 9525 | 9529.19 | P0ACF0_DBHA | hupA | Cytoplasm/Periplasm | 9.57 | 15.88 | 91.22 | -0.228 | NO |
| 10124 | 10131.47 | P0ADZ4_RS15 | rpsO | Cytoplasm | 10.40 | 46.76 | 90.91 | -0.673 | NO |
| 10676 | 10685.76 | P68919_RL25 | rplY | Cytoplasm | 9.60 | 31.89 | 91.28 | -0.392 | YES |
| 11196 | 11192.22 | P0ADZ0_RL23 | rplW | Cytoplasm | 9.94 | 21.46 | 97.40 | -0.373 | YES |
| 12203 | 11208.28 | P60624_RL24 | rplX | Cytoplasm | 10.21 | 9.01 | 89.81 | -0.405 | YES |
| 12638 | 12645.59 | P0AD49_YFIA | raiA | Cytoplasm | 6.18 | 47.48 | 90.62 | -0.535 | YES |
| 12753 | 12761.93 | P0C018_RL18 | rplR | Cytoplasm | 10.42 | 32.35 | 86.92 | -0.395 | YES |
| 12971 | 12960.23 | P0A7S9_RS13 | rpsM | Cytoplasm | 10.78 | 38.74 | 96.75 | -0.424 | YES |
| 13522 | 13518.11 | P0AF93_RIDA | ridA | Cytoplasm | 5.36 | 40.34 | 97.56 | 0.086 | NO |
| 14343 | 14355.66 | P0AG44_RL17 | rplQ | Cytoplasm | 11.05 | 45.90 | 79.29 | -0.565 | NO |
| 14942 | 14957.31 | P02413_RL15 | rplO | Cytoplasm | 11.18 | 43.69 | 87.43 | -0.252 | YES |
| 15385 | 15398.99 | P0ACF8_HNS | hns | Cytoplasm | 5.44 | 40.47 | 82.70 | -0.751 | NO |
| 17547 | 17569.35 | P0A7J3_RL10 | rplJ | Cytoplasm | 9.04 | 28.74 | 94.21 | 0.045 | NO |
| 18745 | 18761.12 | P0AG55_RL6 | rplF | Cytoplasm | 9.71 | 17.04 | 91.93 | -0.227 | NO |
Table 1 Properties of proteins assigned on the MALDI fingerprintsof E. coli
| MALDI(m/z) | TDP precursormass | Protein entry name | Gene ID | Subcellularlocation | Isoelectricpoint | Instability index | Aliphaticindex | GRAVY | Detected by BUP |
|---|---|---|---|---|---|---|---|---|---|
| 6404 | 6406.60 | P0AG51_RL30 | rpmD | Cytoplasm | 10.96 | 38.72 | 102.59 | -0.137 | YES |
| 6849 | 6848.53 | P0AAZ7_YCAR | ycaR | Cytoplasm | 4.31 | 36.22 | 105.39 | -0.127 | YES |
| 7264 | 7268.98 | P0A7M6_RL29 | rpmC | Cytoplasm | 9.98 | 50.27 | 102.22 | -0.657 | YES |
| 7861 | 7865.92 | P0A7M9_RL31 | rpmE | Cytoplasm | 9.46 | 48.68 | 57.00 | -0.649 | NO |
| 8107 | 8113.28 | P69222_IF1 | infA | Cytoplasm | 9.23 | 36.00 | 91.83 | -0.354 | NO |
| 8354 | 8344.10 | P68206_YJBJ | yjbJ | Cytoplasm/Periplasm | 5.64 | 38.38 | 15.60 | -2.432 | NO |
| 8871 | 8869.83 | P0A7M2_RL28 | rpmB | Cytoplasm | 11.42 | 36.21 | 77.87 | -0.650 | YES |
| 9213 | 9219.99 | P0ACF4_DBHB | hupB | Cytoplasm/Periplasm | 9.69 | 16.40 | 92.44 | -0.042 | YES |
| 9525 | 9529.19 | P0ACF0_DBHA | hupA | Cytoplasm/Periplasm | 9.57 | 15.88 | 91.22 | -0.228 | NO |
| 10124 | 10131.47 | P0ADZ4_RS15 | rpsO | Cytoplasm | 10.40 | 46.76 | 90.91 | -0.673 | NO |
| 10676 | 10685.76 | P68919_RL25 | rplY | Cytoplasm | 9.60 | 31.89 | 91.28 | -0.392 | YES |
| 11196 | 11192.22 | P0ADZ0_RL23 | rplW | Cytoplasm | 9.94 | 21.46 | 97.40 | -0.373 | YES |
| 12203 | 11208.28 | P60624_RL24 | rplX | Cytoplasm | 10.21 | 9.01 | 89.81 | -0.405 | YES |
| 12638 | 12645.59 | P0AD49_YFIA | raiA | Cytoplasm | 6.18 | 47.48 | 90.62 | -0.535 | YES |
| 12753 | 12761.93 | P0C018_RL18 | rplR | Cytoplasm | 10.42 | 32.35 | 86.92 | -0.395 | YES |
| 12971 | 12960.23 | P0A7S9_RS13 | rpsM | Cytoplasm | 10.78 | 38.74 | 96.75 | -0.424 | YES |
| 13522 | 13518.11 | P0AF93_RIDA | ridA | Cytoplasm | 5.36 | 40.34 | 97.56 | 0.086 | NO |
| 14343 | 14355.66 | P0AG44_RL17 | rplQ | Cytoplasm | 11.05 | 45.90 | 79.29 | -0.565 | NO |
| 14942 | 14957.31 | P02413_RL15 | rplO | Cytoplasm | 11.18 | 43.69 | 87.43 | -0.252 | YES |
| 15385 | 15398.99 | P0ACF8_HNS | hns | Cytoplasm | 5.44 | 40.47 | 82.70 | -0.751 | NO |
| 17547 | 17569.35 | P0A7J3_RL10 | rplJ | Cytoplasm | 9.04 | 28.74 | 94.21 | 0.045 | NO |
| 18745 | 18761.12 | P0AG55_RL6 | rplF | Cytoplasm | 9.71 | 17.04 | 91.93 | -0.227 | NO |
| MALDI(m/z) | TDP precursormass | Proteinentry name | Gene ID | Subcellularlocation | Isoelectricpoint | Instability index | Aliphaticindex | GRAVY | Detected by BUP |
|---|---|---|---|---|---|---|---|---|---|
| 3342 | 3342.72 | P02968_FLA | hag | Cytoplasm | 6.24 | 27.62 | 82.82 | -0.513 | YES |
| 4376 | 4377.46 | O32107_YUID | yuiD | Membrane | 8.39 | 31.42 | 120.00 | 0.382 | YES |
| 4769 | 4774.53 | P80868_EFG | fusA | Cytoplasm | 8.25 | 48.35 | 76.00 | -0.511 | YES |
| 5042 | 5039.81 | O32111_YUZG | yuzG | Membrane/Secreted | 9.40 | 27.87 | 129.35 | 0.548 | NO |
| 5431 | 5427.16 | P24469_C550 | cccA | Membrane | 6.37 | 19.70 | 77.17 | -0.549 | YES |
| 6099 | 6089.51 | O05522_TATAY | tatAy | Membrane/Secreted | 6.18 | 30.93 | 95.96 | -0.258 | YES |
| 6508 | 6502.85 | P21476_RS19 | rpsS | Cytoplasm/Membrane | 10.11 | 48.28 | 64.57 | -0.740 | YES |
| 6640 | 6641.39 | P23308_SINI | sinI | Cytoplasm/Secreted | 6.26 | 55.83 | 73.68 | -0.804 | NO |
| 6809 | 6807.46 | C0H3Y1_YHZD | yhzD | Cytoplasm/Secreted | 5.79 | 13.70 | 83.28 | -0.461 | NO |
| 7012 | 7009.60 | P70994_4OT | ywhB | Cytoplasm | 5.37 | 59.42 | 75.32 | -0.644 | YES |
| 7116 | 7116.98 | P37815_ATPL | atpE | Membrane | 6.05 | 14.25 | 145.14 | 1.274 | YES |
| 7368 | 7360.61 | P32081_CSPB | cspB | Cytoplasm | 4.54 | 18.56 | 66.87 | -0.337 | YES |
| 7445 | 7438.68 | O03223_RL31 | rpmE | Cytoplasm/Secreted | 9.21 | 35.10 | 47.27 | -0.689 | YES |
| 7715 | 7708.26 | P12873_RL29 | rpmC | Cytoplasm | 10.10 | 34.04 | 96.21 | -0.626 | YES |
| 8255 | 8247.24 | O31718_RPOY | rpoY | Cytoplasm | 4.81 | 50.16 | 83.19 | -0.616 | YES |
| 8546 | 8539.41 | O32119_YUTI | yutI | Cytoplasm | 4.34 | 47.47 | 116.96 | 0.200 | YES |
| 8840 | 8832.90 | P21475_RS18 | rpsR | Cytoplasm/Secreted | 11.04 | 43.37 | 76.58 | -0.639 | YES |
| 9209 | 9201.90 | P05657_RL27 | rpmA | Membrane/Secreted | 10.32 | 11.28 | 57.79 | -0.809 | YES |
| 9470 | 9462.29 | P21477_RS20 | rpsT | Cytoplasm/Secreted | 10.94 | 30.28 | 77.84 | -0.714 | YES |
| 9886 | 9878.28 | P08821_DBH1 | hupA | Cytoplasm | 8.96 | 29.52 | 82.83 | -0.485 | YES |
| 10451 | 10445.56 | P21476_RS19 | rpsS | Cytoplasm/Membrane | 10.11 | 48.28 | 64.57 | -0.740 | YES |
| 10952 | 10943.59 | P28015_SP5G | spoVG | Cytoplasm | 5.25 | 26.40 | 83.40 | -0.479 | YES |
| 11150 | 11163.36 | P0CI78_RL24 | rplX | Cytoplasm/Membrane | 10.04 | 16.40 | 77.38 | -0.644 | YES |
| MALDI(m/z) | TDP precursormass | Proteinentry name | Gene ID | Subcellularlocation | Isoelectricpoint | Instability index | Aliphaticindex | GRAVY | Detected by BUP |
| 11536 | 11527.33 | P21471_RS10 | rpsJ | Cytoplasm | 9.79 | 48.41 | 99.41 | -0.460 | YES |
| 11591 | 11585.06 | P0C174_GPSB | gpsB | Cytoplasm | 5.72 | 50.84 | 85.51 | -0.853 | YES |
| 12463 | 12451.83 | P42060_RL22 | rplV | Cytoplasm | 10.73 | 31.93 | 97.61 | -0.244 | YES |
| 12622 | 12611.85 | P02394_RL7 | rplL | Cytoplasm | 4.56 | 44.34 | 114.31 | 0.115 | YES |
| 12972 | 12960.98 | P46899_RL18 | rplR | Cytoplasm | 10.11 | 19.12 | 87.92 | -0.370 | YES |
| 13157 | 13146.12 | P12875_RL14 | rplN | Cytoplasm | 9.94 | 31.75 | 96.56 | -0.137 | YES |
| 13390 | 13378.60 | O31742_RL19 | rplS | Cytoplasm | 10.97 | 41.84 | 98.17 | -0.475 | YES |
| 13652 | 13647.71 | P20282_RS13 | rpsM | Cytoplasm | 11.07 | 42.95 | 96.61 | -0.720 | YES |
| 14213 | 14210.74 | P21470_RS9 | rpsI | Cytoplasm/Membrane | 10.60 | 40.42 | 87.85 | -0.512 | YES |
| 15286 | 15271.55 | P35137_PPIB | ppiB | Cytoplasm | 5.53 | 20.21 | 59.30 | -0.364 | YES |
| 16380 | 16364.70 | P70974_RL13 | rplM | Cytoplasm | 9.87 | 15.64 | 80.07 | -0.632 | YES |
| 17631 | 17611.78 | P21467_RS5 | rpsE | Cytoplasm | 9.92 | 34.39 | 108.61 | -0.001 | YES |
| 17770 | 17754.49 | P21469_RS7 | rpsG | Cytoplasm | 10.81 | 45.66 | 98.17 | -0.621 | YES |
| 19385 | 19366.40 | P46898_RL6 | rplF | Cytoplasm | 9.49 | 31.26 | 87.49 | -0.501 | YES |
Table 2 Properties of proteins assigned on the MALDI fingerprintsof B. subtilis
| MALDI(m/z) | TDP precursormass | Proteinentry name | Gene ID | Subcellularlocation | Isoelectricpoint | Instability index | Aliphaticindex | GRAVY | Detected by BUP |
|---|---|---|---|---|---|---|---|---|---|
| 3342 | 3342.72 | P02968_FLA | hag | Cytoplasm | 6.24 | 27.62 | 82.82 | -0.513 | YES |
| 4376 | 4377.46 | O32107_YUID | yuiD | Membrane | 8.39 | 31.42 | 120.00 | 0.382 | YES |
| 4769 | 4774.53 | P80868_EFG | fusA | Cytoplasm | 8.25 | 48.35 | 76.00 | -0.511 | YES |
| 5042 | 5039.81 | O32111_YUZG | yuzG | Membrane/Secreted | 9.40 | 27.87 | 129.35 | 0.548 | NO |
| 5431 | 5427.16 | P24469_C550 | cccA | Membrane | 6.37 | 19.70 | 77.17 | -0.549 | YES |
| 6099 | 6089.51 | O05522_TATAY | tatAy | Membrane/Secreted | 6.18 | 30.93 | 95.96 | -0.258 | YES |
| 6508 | 6502.85 | P21476_RS19 | rpsS | Cytoplasm/Membrane | 10.11 | 48.28 | 64.57 | -0.740 | YES |
| 6640 | 6641.39 | P23308_SINI | sinI | Cytoplasm/Secreted | 6.26 | 55.83 | 73.68 | -0.804 | NO |
| 6809 | 6807.46 | C0H3Y1_YHZD | yhzD | Cytoplasm/Secreted | 5.79 | 13.70 | 83.28 | -0.461 | NO |
| 7012 | 7009.60 | P70994_4OT | ywhB | Cytoplasm | 5.37 | 59.42 | 75.32 | -0.644 | YES |
| 7116 | 7116.98 | P37815_ATPL | atpE | Membrane | 6.05 | 14.25 | 145.14 | 1.274 | YES |
| 7368 | 7360.61 | P32081_CSPB | cspB | Cytoplasm | 4.54 | 18.56 | 66.87 | -0.337 | YES |
| 7445 | 7438.68 | O03223_RL31 | rpmE | Cytoplasm/Secreted | 9.21 | 35.10 | 47.27 | -0.689 | YES |
| 7715 | 7708.26 | P12873_RL29 | rpmC | Cytoplasm | 10.10 | 34.04 | 96.21 | -0.626 | YES |
| 8255 | 8247.24 | O31718_RPOY | rpoY | Cytoplasm | 4.81 | 50.16 | 83.19 | -0.616 | YES |
| 8546 | 8539.41 | O32119_YUTI | yutI | Cytoplasm | 4.34 | 47.47 | 116.96 | 0.200 | YES |
| 8840 | 8832.90 | P21475_RS18 | rpsR | Cytoplasm/Secreted | 11.04 | 43.37 | 76.58 | -0.639 | YES |
| 9209 | 9201.90 | P05657_RL27 | rpmA | Membrane/Secreted | 10.32 | 11.28 | 57.79 | -0.809 | YES |
| 9470 | 9462.29 | P21477_RS20 | rpsT | Cytoplasm/Secreted | 10.94 | 30.28 | 77.84 | -0.714 | YES |
| 9886 | 9878.28 | P08821_DBH1 | hupA | Cytoplasm | 8.96 | 29.52 | 82.83 | -0.485 | YES |
| 10451 | 10445.56 | P21476_RS19 | rpsS | Cytoplasm/Membrane | 10.11 | 48.28 | 64.57 | -0.740 | YES |
| 10952 | 10943.59 | P28015_SP5G | spoVG | Cytoplasm | 5.25 | 26.40 | 83.40 | -0.479 | YES |
| 11150 | 11163.36 | P0CI78_RL24 | rplX | Cytoplasm/Membrane | 10.04 | 16.40 | 77.38 | -0.644 | YES |
| MALDI(m/z) | TDP precursormass | Proteinentry name | Gene ID | Subcellularlocation | Isoelectricpoint | Instability index | Aliphaticindex | GRAVY | Detected by BUP |
| 11536 | 11527.33 | P21471_RS10 | rpsJ | Cytoplasm | 9.79 | 48.41 | 99.41 | -0.460 | YES |
| 11591 | 11585.06 | P0C174_GPSB | gpsB | Cytoplasm | 5.72 | 50.84 | 85.51 | -0.853 | YES |
| 12463 | 12451.83 | P42060_RL22 | rplV | Cytoplasm | 10.73 | 31.93 | 97.61 | -0.244 | YES |
| 12622 | 12611.85 | P02394_RL7 | rplL | Cytoplasm | 4.56 | 44.34 | 114.31 | 0.115 | YES |
| 12972 | 12960.98 | P46899_RL18 | rplR | Cytoplasm | 10.11 | 19.12 | 87.92 | -0.370 | YES |
| 13157 | 13146.12 | P12875_RL14 | rplN | Cytoplasm | 9.94 | 31.75 | 96.56 | -0.137 | YES |
| 13390 | 13378.60 | O31742_RL19 | rplS | Cytoplasm | 10.97 | 41.84 | 98.17 | -0.475 | YES |
| 13652 | 13647.71 | P20282_RS13 | rpsM | Cytoplasm | 11.07 | 42.95 | 96.61 | -0.720 | YES |
| 14213 | 14210.74 | P21470_RS9 | rpsI | Cytoplasm/Membrane | 10.60 | 40.42 | 87.85 | -0.512 | YES |
| 15286 | 15271.55 | P35137_PPIB | ppiB | Cytoplasm | 5.53 | 20.21 | 59.30 | -0.364 | YES |
| 16380 | 16364.70 | P70974_RL13 | rplM | Cytoplasm | 9.87 | 15.64 | 80.07 | -0.632 | YES |
| 17631 | 17611.78 | P21467_RS5 | rpsE | Cytoplasm | 9.92 | 34.39 | 108.61 | -0.001 | YES |
| 17770 | 17754.49 | P21469_RS7 | rpsG | Cytoplasm | 10.81 | 45.66 | 98.17 | -0.621 | YES |
| 19385 | 19366.40 | P46898_RL6 | rplF | Cytoplasm | 9.49 | 31.26 | 87.49 | -0.501 | YES |
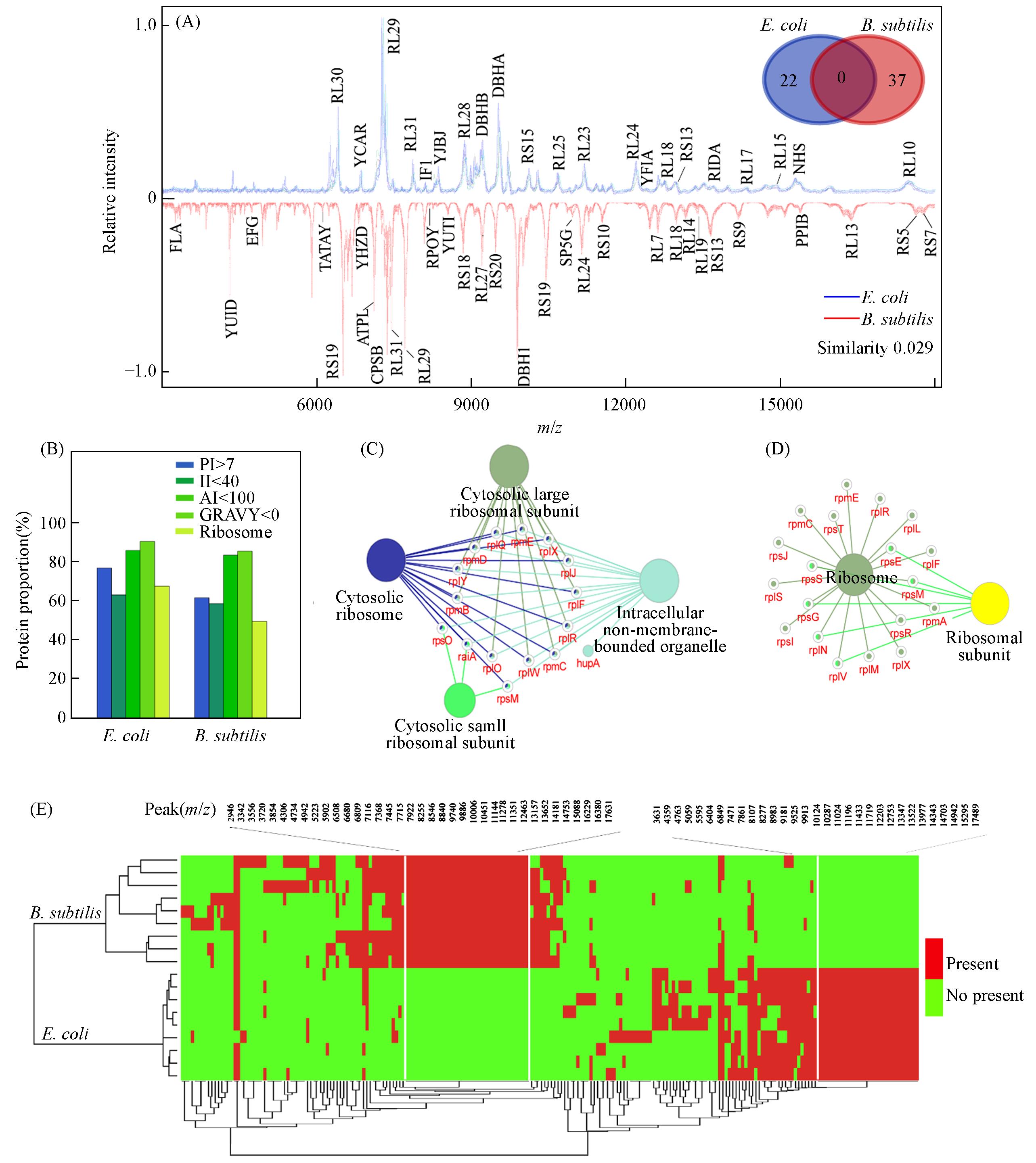
Fig.3 Correlation between MALDI and TDP protein profiles(A) MALDI protein fingerprints of E. coli and B. subtilis, with protein identity labeled on the main peaks. The protein identity was tentatively assigned through matching the fingerprint peak masses to the protein(or protein fragment) masses measured by TDP; insert: fingerprint peak numbers detected from E. coli and B. subtilis with protein identity assigned; (B) investigation of the properties of proteins assigned on the MALDI fingerprints of E. coli and B. subtilis. The investigated properties include isoelectric point(PI), instability index(II), aliphatic index(AI), grand average of hydropathy(GRAVY), and if the proteins were ribosomal proteins; (C) biological functional analysis of the proteins assigned for E. coli using ClueGO system; (D) biological functional analysis of the proteins assigned for B. subtilis using ClueGO system; gene ontology(GO) terms are represented as nodes, with node size representing the enrichment significance; only the most significant terms were labeled, with the exhibition of representative enriched pathway(P < 0.05) interactions among the main targets; (E) a deep hierarchical clustering analysis of the MALDI fingerprints using cosine correlation distance function and average linkage, correlated with the peak presence profile. The peaks exclusively detected from E. coli and B. subtilis were listed.
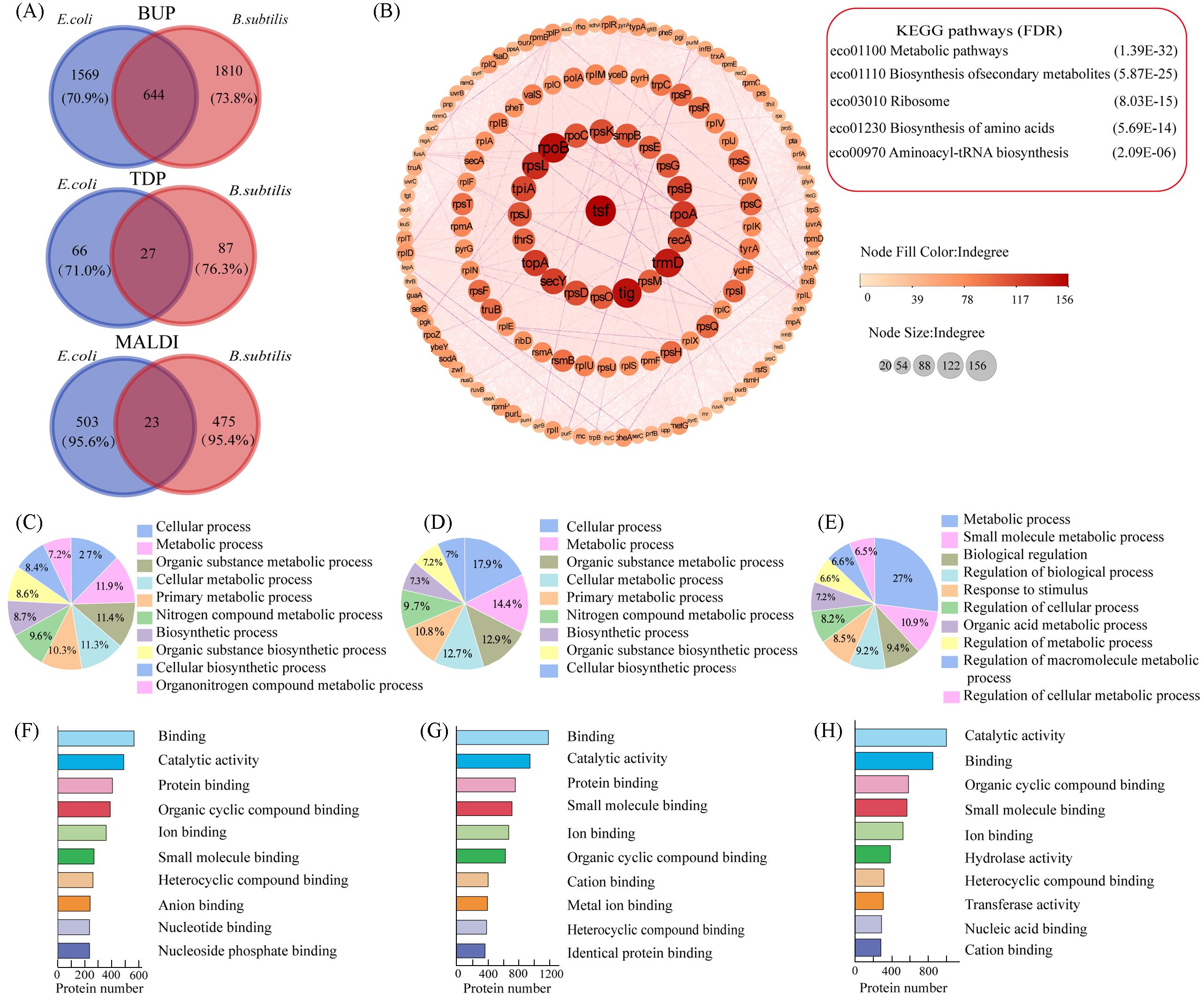
Fig.5 BUP profile exhibited the largest overlap between bacterial species(A) Counts of species-common and species-specific proteins or protein peaks detected from E. coli and B. subtilis using MALDI, TDP and BUP respectively; (B) the protein-protein interaction network of the BUP species-common proteins, constructed using the String database and Cytoscape with an indegree score threshold of 30; (C—E) top ten significantly enriched biological processes(BPs) from the species-common proteins(C), E. coli-specific proteins(D) and B. subtilis-specific proteins(E) detected by BUP; (F—H) top ten significantly enriched molecular functions(MFs) from the species-common proteins(F), E. coli-specific proteins(G) and B. subtilis-specific proteins(H) detected by BUP. P-value lower than 0.05 indicated a significant enrichment.
| 1 | Zhao X., Bi H. Y., J. Agric. Food Chem., 2022, 70(24), 7525—7534 |
| 2 | Yang K. B., Cameranesi M., Gowder M., Martinez C., Shamovsky Y., Epshtein V., Hao Z. T., Nguyen T., Nirenstein E., Shamovsky I., Rasouly A., Nudler E., Nature, 2023, 622(7981), 180—187 |
| 3 | Chirania P., Holwerda E. K., Giannone R. J., Liang X. Y., Poudel S., Ellis J. C., Bomble Y. J., Hettich R. L., Lynd L. R., Nat. Commun., 2022, 13, 3870 |
| 4 | Schwenzer A. K., Kruse L., Jooss K., Neususs C., Proteomics, 2024, 24(3-4), e2300135 |
| 5 | Zhou F., Johnston M. V., Electrophoresis, 2005, 26(7/8), 1383—1388 |
| 6 | Lee P. Y., Saraygord⁃Afshari N., Low T. Y., J. Chromatogr. A, 2020, 1615, 460763 |
| 7 | Rebecca W., Scheller C., Krebs F., Wätzig H., Oltmann‐Norden I., Electrophoresis, 2020, 42(3), 206—218 |
| 8 | Zhu Y. D., Qiao L., Prudent M., Bondarenko A., Gasilova N., Möller S. B., Lion N., Pick H., Gong T. Q., Chen Z. X., Yang P. Y., Lovey L. T., Girault H. H., Chem. Sci., 2016, 7(5), 2987—2995 |
| 9 | Toby T. K., Fornelli L., Srzentic K., DeHart C. J., Levitsky J., Friedewald J., Kelleher N. L., Nat. Protoc., 2019, 14(1), 119—152 |
| 10 | Dupree E. J., Jayathirtha M., Yorkey H., Mihasan M., Petre B. A., Darie C. C., Proteomes, 2020, 8(3), 8030014 |
| 11 | Gibson B., Wilson D. J., Feil E., Eyre⁃Walker A., Proc. Biol. Sci., 2018, 285(1880), 20180789 |
| 12 | Du M. G., Yuan Z. N., Werneburg G. T., Henderson N. S., Chauhan H., Kovach A., Zhao G. P., Johl J., Li H. L., Thanassi D. G., Nat. Commun., 2021, 12(1), 5207 |
| 13 | Nefedov A. V., Mitra I., Brasier A. R., Sadygov R. G., J. Proteome Res., 2011, 10(9), 4150—4157 |
| 14 | van Doorn J., Ly A., Marsman M., Wagenmakers E. J., J. Appl. Statist., 2020, 47(16), 2984—3006 |
| 15 | Jaskolla T. W., Karas M., J. Am. Soc. Mass Spectrom., 2011, 22(6), 976—988 |
| 16 | Zhu Y. D., Pick H., Gasilova N., Li X. Y., Lin T. E., Laeubli H. P., Zippelius A., Ho P. C., Girault H. H., Chem, 2019, 5(5), 1318—1336 |
| 17 | Yu C. S., Lin C. J., Hwang J. K., Prot. Sci., 2004, 13(5), 1402—1406 |
| 18 | Bindea G., Mlecnik B., Hackl H., Charoentong P., Tosolini M., Kirilovsky A., Fridman W. H., Pagès F., Trajanoski Z., Galon J., Bioinformatics, 2009, 25(8), 1091—1093 |
| 19 | Fromm S. A., O'Connor K. M., Purdy M., Bhatt P. R., Loughran G., Atkins J. F., Jomaa A., Mattei S., Nat. Commun., 2023, 14(1), 1095 |
| 20 | Ishihama Y., Schmidt T., Rappsilber J., Mann M., Hartl F. U., Kerner M. J., Frishman D., BMC Genom., 2008, 9, 102 |
| 21 | Zundel M. A., Basturea G. N., Deutscher M. P., RNA, 2009, 15(5), 977—983 |
| 22 | Fedyukina D. V., Jennaro T. S., Cavagnero S., J. Biol. Chem., 2014, 289(10), 6740—6750 |
| 23 | Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., Ideker T., Genom. Res., 2003, 13(11), 2498—2504 |
| 24 | Burnett B. J., Altman R. B., Ferrao R., Alejo J. L., Kaur N., Kanji J., Blanchard S. C., J. Biol. Chem., 2013, 288(19), 13917—13928 |
| 25 | Houry W., Protein Sci., 2021, 30, 24—25 |
| 26 | Rungsirivanich P., Inta A., Tragoolpua Y., Thongwai N., Sci. Rep., 2019, 9, 16561 |
| 27 | de Crécy⁃Lagard V., Ross R. L., Jaroch M., Marchand V., Eisenhart C., Brégeon D., Motorin Y., Limbach P. A., Biomolecules, 2020, 10(7), 977 |
| [1] | YANG Jiaqiang, WU Xuejiao, LU Zicong, CHEN Yangmi, SHE Huixian, LIU Ouling. Synthesis and Antibacterial Activities of Osthole Derivatives Containing (Hetero)arylsulfonylpiperazine [J]. Chem. J. Chinese Universities, 2024, 45(7): 20240124. |
| [2] | ZHOU Xiaolong, WANG Ruiqi, CHEN Guoli, GUO Wenming, CHEN Fei, ZHOU Yan, SUN Lide, TANG Dabao, XU Yunhui. Synthesis of O-quaternary Ammonium Salt-oxidized Chitosan and Its Antibacterial Finishing for Cotton Fabric [J]. Chem. J. Chinese Universities, 2024, 45(3): 20230375. |
| [3] | SHI Qian, LIU Dongmei, FANG Xiaoni, LIU Baohong. High-throughput Analysis of Tyrosinase Activity and Inhibitors Based on Matrix-assisted Laser Desorption/ionization Mass Spectrometry [J]. Chem. J. Chinese Universities, 2024, 45(11): 20240330. |
| [4] | YAN Yongjie, GAO Wenbo, LU Chenhui, YANG Cheng, XU Shuting. Detection of Coffee Polyphenols in Nanoliter Cerebrospinal Fluid by Microextraction Coupled to Nanoelectrospray Ionization Mass Spectrometry [J]. Chem. J. Chinese Universities, 2024, 45(11): 20240327. |
| [5] | ZHANG Lei, SHEN Huali. Quantitative Accuracy Evaluation of Mass Spectrometry Based Proteomics Methods Commonly Used in Biomarker Research [J]. Chem. J. Chinese Universities, 2024, 45(11): 20240311. |
| [6] | JIN Ying, ZHANG Junjie, ZHANG Yixin, YUAN Yue, HAN Zhenzhen. Research Progress in Exosome Isolation and Proteomics Analysis [J]. Chem. J. Chinese Universities, 2024, 45(11): 20240305. |
| [7] | HU Yuhong, YU Xiangming, SONG Lili, XING Qinghe, ZHOU Feng. Dynamic Proteome Profiling of Neonatal Hair Shaft Using DEEP SEQ Method [J]. Chem. J. Chinese Universities, 2024, 45(11): 20240326. |
| [8] | FAN Zhirui, FANG Qun, YANG Yi. Single Cell Proteomic Analysis by Mass Spectrometry [J]. Chem. J. Chinese Universities, 2024, 45(11): 20240294. |
| [9] | XU Xia, QIN Weida, LI Ruomeng, WANG Qianqian, LIU Ning, LI Gongyu. Mass Spectrometry-based Deep Coverage Proteome: Evaluation of Cellular Protein Extraction Methods [J]. Chem. J. Chinese Universities, 2024, 45(11): 20240344. |
| [10] | XU Hongmei, WANG Liangchen, MIN Qianhao. Advances in MALDI MS Matrices for the Detection of Small Molecules [J]. Chem. J. Chinese Universities, 2024, 45(11): 20240285. |
| [11] | LIU Siying, SU Wen, ZHOU Zhongyan, YANG Zhiyu, PEI Huafu, HE Zhiru, WANG Na, YUE Lei. Protein-Small Molecule Interaction Electrospray Ionization Mass Spectrometry Study of the Ubiquitin/Adenosine Triphoshate Couple over Temperature Variation [J]. Chem. J. Chinese Universities, 2024, 45(11): 20240382. |
| [12] | SHEN Fenglin, FENG Zhaoying, FANG Jing, ZHANG Lei, LIU Xiaohui, ZHOU Xinwen. New Technologies for Mass Spectrometry-based Single-cell Resolved Spatial Proteomics Research [J]. Chem. J. Chinese Universities, 2024, 45(11): 20240299. |
| [13] | HUO Zhiyuan, ZHOU Jinping, MA Xiumin, ZHOU Yan, HUANG Lin. Advances in Single-cell Multi-omics Analysis Based on Mass Spectrometry [J]. Chem. J. Chinese Universities, 2024, 45(11): 20240389. |
| [14] | DONG Peiying, LIU Tong, QIN Weijie. A New Method for Large-scale Enrichment and Stepwise Identification of RNA-protein Complexes [J]. Chem. J. Chinese Universities, 2024, 45(11): 20240091. |
| [15] | CAO Ting, SHU Weikang, WAN Jingjing. Plasmonic Composites Aid Semi-quantitative Analysis and Identification of Low-molecular-weight Metabolites by Mass Spectrometry [J]. Chem. J. Chinese Universities, 2024, 45(11): 20240325. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||