

Chem. J. Chinese Universities ›› 2019, Vol. 40 ›› Issue (6): 1141.doi: 10.7503/cjcu20180782
• Analytical Chemistry • Previous Articles Next Articles
WEI Wanghui1, CHU Yanqiu2( ), CHEN Ying1, Gao Yanqiu1, DING Chuanfan2(
), CHEN Ying1, Gao Yanqiu1, DING Chuanfan2( )
)
Received:2018-11-21
Online:2019-06-10
Published:2019-04-04
Supported by:CLC Number:
TrendMD:
WEI Wanghui,CHU Yanqiu,CHEN Ying,Gao Yanqiu,DING Chuanfan. Quantitative Determination of the Glycosylation Level for the Binding Proteins to High Density Lipoprotein by Mass Spectrometry†[J]. Chem. J. Chinese Universities, 2019, 40(6): 1141.
| No. | Protein | Peptidesequence | Site | Glycan moiety | MS(calcd.), m/z | Retention time/min | |
|---|---|---|---|---|---|---|---|
| Precursor ion | Product ion | ||||||
| 1 | SAA | GPGGVWAAEAISDAR | 486.4 | 448.2, 561.3 | 6.82 | ||
| 2 | A1AT | AVLTIDEK | 444.8 | 718.4, 605.3 | 4.99 | ||
| 3 | A1AT | QLAHQSNSTNIFFSPVSIATAFAMLSLGTK | 70 | 5_4_0_2 | 1078.5 | 366.1 | 10.58 |
| 4 | A1AT | QLAHQSNSTNIFFSPVSIATAFAMLSLGTK | 70 | 5_4_1_2 | 1107.7 | 366.1 | 10.29 |
| 5 | A1AT | ADTHDEILEGLNFNLTEIPEAQIHEGFQELLR | 107 | 5_4_1_1 | 1151.6 | 366.1 | 9.15 |
| 6 | A1AT | ADTHDEILEGLNFNLTEIPEAQIHEGFQELLR | 107 | 6_5_0_3 | 1311.8 | 366.1 | 9.43 |
| 7 | A1AT | ADTHDEILEGLNFNLTEIPEAQIHEGFQELLR | 107 | 6_5_1_3 | 1341.0 | 366.1 | 9.43 |
| 8 | A1AT | YLGNATAIFFLPDEGK | 271 | 5_4_0_2 | 991.2 | 366.1 | 8.23 |
| 9 | A1AT | YLGNATAIFFLPDEGK | 271 | 5_4_1_2 | 1027.7 | 366.1 | 6.74 |
Table 1 MRM transitions for monitoring peptide of SAA and peptide, glycopeptides of A1AT
| No. | Protein | Peptidesequence | Site | Glycan moiety | MS(calcd.), m/z | Retention time/min | |
|---|---|---|---|---|---|---|---|
| Precursor ion | Product ion | ||||||
| 1 | SAA | GPGGVWAAEAISDAR | 486.4 | 448.2, 561.3 | 6.82 | ||
| 2 | A1AT | AVLTIDEK | 444.8 | 718.4, 605.3 | 4.99 | ||
| 3 | A1AT | QLAHQSNSTNIFFSPVSIATAFAMLSLGTK | 70 | 5_4_0_2 | 1078.5 | 366.1 | 10.58 |
| 4 | A1AT | QLAHQSNSTNIFFSPVSIATAFAMLSLGTK | 70 | 5_4_1_2 | 1107.7 | 366.1 | 10.29 |
| 5 | A1AT | ADTHDEILEGLNFNLTEIPEAQIHEGFQELLR | 107 | 5_4_1_1 | 1151.6 | 366.1 | 9.15 |
| 6 | A1AT | ADTHDEILEGLNFNLTEIPEAQIHEGFQELLR | 107 | 6_5_0_3 | 1311.8 | 366.1 | 9.43 |
| 7 | A1AT | ADTHDEILEGLNFNLTEIPEAQIHEGFQELLR | 107 | 6_5_1_3 | 1341.0 | 366.1 | 9.43 |
| 8 | A1AT | YLGNATAIFFLPDEGK | 271 | 5_4_0_2 | 991.2 | 366.1 | 8.23 |
| 9 | A1AT | YLGNATAIFFLPDEGK | 271 | 5_4_1_2 | 1027.7 | 366.1 | 6.74 |
| No. | Protein | Peptidesequence | Site | Glycan moiety | MS(calcd.), m/z | Retention time/min | |
|---|---|---|---|---|---|---|---|
| Precursor ion | Product ion | ||||||
| 1 | A2HSG | EATEAAK | 360.2 | 289.1, 519.3 | 2.84 | ||
| 2 | A2HSG | VCQDCPLLAPLNDTR | 156 | 5_4_0_1 | 1191.2 | 366.1 | 6.87 |
| 3 | A2HSG | VCQDCPLLAPLNDTR | 156 | 5_4_0_2 | 1288.2 | 366.1 | 7.03 |
| 4 | A2HSG | VCQDCPLLAPLNDTR | 156 | 5_4_1_2 | 1002.9 | 366.1 | 6.99 |
| 5 | A2HSG | VCQDCPLLAPLNDTR | 156 | 5_4_2_1 | 966.6 | 366.1 | 7.01 |
| 6 | A2HSG | VCQDCPLLAPLNDTR | 156 | 6_5_0_2 | 1057.7 | 366.1 | 6.99 |
| 7 | A2HSG | VCQDCPLLAPLNDTR | 156 | 6_5_0_3 | 1130.5 | 366.1 | 7.08 |
| 8 | A2HSG | VCQDCPLLAPLNDTR | 156 | 6_5_1_0 | 948.6 | 366.1 | 7.17 |
| 9 | A2HSG | VCQDCPLLAPLNDTR | 156 | 6_5_1_3 | 1167.0 | 366.1 | 7.06 |
| 10 | A2HSG | AALAAFNAQNNGSNFQLEEISR | 176 | 5_4_0_1 | 1070.4 | 366.1 | 7.11 |
| 11 | A2HSG | AALAAFNAQNNGSNFQLEEISR | 176 | 5_4_0_2 | 1143.0 | 366.1 | 7.22 |
| 12 | A2HSG | AALAAFNAQNNGSNFQLEEISR | 176 | 5_4_1_2 | 1179.7 | 366.1 | 7.2 |
| 13 | A2HSG | AALAAFNAQNNGSNFQLEEISR | 176 | 5_4_3_1 | 1180.3 | 366.1 | 7.2 |
| 14 | A2HSG | AALAAFNAQNNGSNFQLEEISR | 176 | 6_5_0_1 | 1161.7 | 366.1 | 7.09 |
| 15 | A2HSG | AALAAFNAQNNGSNFQLEEISR | 176 | 6_5_0_2 | 1234.3 | 366.1 | 7.17 |
| 16 | A2HSG | AALAAFNAQNNGSNFQLEEISR | 176 | 6_5_0_3 | 1307.1 | 366.1 | 7.3 |
| No. | Protein | Peptidesequence | Site | Glycan moiety | MS(calcd.), m/z | Retention time/min | |
| Precursor ion | Product ion | ||||||
| 17 | A2HSG | AALAAFNAQNNGSNFQLEEISR | 176 | 6_5_1_2 | 1271.0 | 366.1 | 7.17 |
| 18 | A2HSG | AALAAFNAQNNGSNFQLEEISR | 176 | 6_5_1_3 | 1343.8 | 366.1 | 7.3 |
| 19 | A2HSG | AALAAFNAQNNGSNFQLEEISR | 176 | 7_6_0_0 | 1180.5 | 366.1 | 7.2 |
| 20 | A2HSG | HTFMGVVSLGSPSGEVSHPR | 319 | 1_1_0_1 | 913.1 | 274.1 | 11.02 |
| 21 | A2HSG | HTFMGVVSLGSPSGEVSHPR | 319 | 1_1_0_2 | 757.8 | 274.1 | 7.89 |
| 22 | A2HSG | HTFMGVVSLGSPSGEVSHPR | 319 | 1_1_1_1 | 961.8 | 274.1 | 11.06 |
| 23 | A2HSG | HTFMGVVSLGSPSGEVSHPR | 319 | 1_2_0_1 | 735.8 | 274.1 | 2.69 |
| 24 | A2HSG | TVVQPSVGAAAGPVVPPCPGR | 346 | 1_1_0_1 | 891.4 | 274.1 | 6.32 |
| 25 | A2HSG | TVVQPSVGAAAGPVVPPCPGR | 346 | 1_1_0_2 | 988.5 | 274.1 | 6.34 |
| 26 | A2HSG | TVVQPSVGAAAGPVVPPCPGR | 346 | 2_1_1_0 | 897.1 | 366.1 | 11.08 |
| 27 | A2HSG | TVVQPSVGAAAGPVVPPCPGR | 346 | 2_2_0_0 | 916.1 | 366.1 | 7.21 |
Table 2 MRM transitions for monitoring peptide and glycopeptides of A2HSG
| No. | Protein | Peptidesequence | Site | Glycan moiety | MS(calcd.), m/z | Retention time/min | |
|---|---|---|---|---|---|---|---|
| Precursor ion | Product ion | ||||||
| 1 | A2HSG | EATEAAK | 360.2 | 289.1, 519.3 | 2.84 | ||
| 2 | A2HSG | VCQDCPLLAPLNDTR | 156 | 5_4_0_1 | 1191.2 | 366.1 | 6.87 |
| 3 | A2HSG | VCQDCPLLAPLNDTR | 156 | 5_4_0_2 | 1288.2 | 366.1 | 7.03 |
| 4 | A2HSG | VCQDCPLLAPLNDTR | 156 | 5_4_1_2 | 1002.9 | 366.1 | 6.99 |
| 5 | A2HSG | VCQDCPLLAPLNDTR | 156 | 5_4_2_1 | 966.6 | 366.1 | 7.01 |
| 6 | A2HSG | VCQDCPLLAPLNDTR | 156 | 6_5_0_2 | 1057.7 | 366.1 | 6.99 |
| 7 | A2HSG | VCQDCPLLAPLNDTR | 156 | 6_5_0_3 | 1130.5 | 366.1 | 7.08 |
| 8 | A2HSG | VCQDCPLLAPLNDTR | 156 | 6_5_1_0 | 948.6 | 366.1 | 7.17 |
| 9 | A2HSG | VCQDCPLLAPLNDTR | 156 | 6_5_1_3 | 1167.0 | 366.1 | 7.06 |
| 10 | A2HSG | AALAAFNAQNNGSNFQLEEISR | 176 | 5_4_0_1 | 1070.4 | 366.1 | 7.11 |
| 11 | A2HSG | AALAAFNAQNNGSNFQLEEISR | 176 | 5_4_0_2 | 1143.0 | 366.1 | 7.22 |
| 12 | A2HSG | AALAAFNAQNNGSNFQLEEISR | 176 | 5_4_1_2 | 1179.7 | 366.1 | 7.2 |
| 13 | A2HSG | AALAAFNAQNNGSNFQLEEISR | 176 | 5_4_3_1 | 1180.3 | 366.1 | 7.2 |
| 14 | A2HSG | AALAAFNAQNNGSNFQLEEISR | 176 | 6_5_0_1 | 1161.7 | 366.1 | 7.09 |
| 15 | A2HSG | AALAAFNAQNNGSNFQLEEISR | 176 | 6_5_0_2 | 1234.3 | 366.1 | 7.17 |
| 16 | A2HSG | AALAAFNAQNNGSNFQLEEISR | 176 | 6_5_0_3 | 1307.1 | 366.1 | 7.3 |
| No. | Protein | Peptidesequence | Site | Glycan moiety | MS(calcd.), m/z | Retention time/min | |
| Precursor ion | Product ion | ||||||
| 17 | A2HSG | AALAAFNAQNNGSNFQLEEISR | 176 | 6_5_1_2 | 1271.0 | 366.1 | 7.17 |
| 18 | A2HSG | AALAAFNAQNNGSNFQLEEISR | 176 | 6_5_1_3 | 1343.8 | 366.1 | 7.3 |
| 19 | A2HSG | AALAAFNAQNNGSNFQLEEISR | 176 | 7_6_0_0 | 1180.5 | 366.1 | 7.2 |
| 20 | A2HSG | HTFMGVVSLGSPSGEVSHPR | 319 | 1_1_0_1 | 913.1 | 274.1 | 11.02 |
| 21 | A2HSG | HTFMGVVSLGSPSGEVSHPR | 319 | 1_1_0_2 | 757.8 | 274.1 | 7.89 |
| 22 | A2HSG | HTFMGVVSLGSPSGEVSHPR | 319 | 1_1_1_1 | 961.8 | 274.1 | 11.06 |
| 23 | A2HSG | HTFMGVVSLGSPSGEVSHPR | 319 | 1_2_0_1 | 735.8 | 274.1 | 2.69 |
| 24 | A2HSG | TVVQPSVGAAAGPVVPPCPGR | 346 | 1_1_0_1 | 891.4 | 274.1 | 6.32 |
| 25 | A2HSG | TVVQPSVGAAAGPVVPPCPGR | 346 | 1_1_0_2 | 988.5 | 274.1 | 6.34 |
| 26 | A2HSG | TVVQPSVGAAAGPVVPPCPGR | 346 | 2_1_1_0 | 897.1 | 366.1 | 11.08 |
| 27 | A2HSG | TVVQPSVGAAAGPVVPPCPGR | 346 | 2_2_0_0 | 916.1 | 366.1 | 7.21 |
| No. | Protein | Peptidesequence | Site | Glycan moiety | MS(calcd.), m/z | Retention time/min | |
|---|---|---|---|---|---|---|---|
| Precursor ion | Product ion | ||||||
| 1 | Apo C3 | GWVTDGFSSLK | 598.8 | 244.2, 854.4 | 7.25 | ||
| 2 | Apo C3 | PTSAVAA | 94 | 0_3_0_0 | 613.3 | 204.1 | 7.21 |
| 3 | Apo C3 | PTSAVAA | 94 | 0_3_1_0 | 686.3 | 204.1 | 11.07 |
| 4 | Apo C3 | PTSAVAA | 94 | 1_1_0_1 | 636.8 | 274.1 | 8.18 |
| 5 | Apo C3 | PTSAVAA | 94 | 1_1_0_2 | 782.3 | 274.1 | 8.29 |
| 6 | Apo C3 | PTSAVAA | 94 | 1_1_1_1 | 709.8 | 274.1 | 7.89 |
| 7 | Apo C3 | PTSAVAA | 94 | 1_2_0_2 | 883.9 | 274.1 | 7.84 |
| 8 | Apo C3 | PTSAVAA | 94 | 1_2_1_0 | 665.8 | 366.1 | 11.08 |
| 9 | Apo C3 | PTSAVAA | 94 | 1_3_0_0 | 694.3 | 366.1 | 11.07 |
| 10 | Apo C3 | PTSAVAA | 94 | 1_3_1_1 | 912.9 | 274.1 | 7.86 |
| 11 | Apo C3 | PTSAVAA | 94 | 2_1_1_0 | 645.3 | 366.1 | 11.07 |
| 12 | Apo C3 | PTSAVAA | 94 | 2_2_0_0 | 673.8 | 366.1 | 7.86 |
| 13 | Apo C3 | PTSAVAA | 94 | 2_2_1_2 | 1037.9 | 274.1 | 6.32 |
| 14 | Apo C3 | PTSAVAA | 94 | 2_2_2_0 | 673.8 | 274.1 | 7.03 |
| 15 | Apo C3 | PTSAVAA | 94 | 2_2_3_0 | 892.9 | 366.1 | 10.58 |
| 16 | Apo C3 | PTSAVAA | 94 | 2_3_0_1 | 920.9 | 274.1 | 7.60 |
Table 3 MRM transitions for monitoring peptide and glycopeptides of Apo C3
| No. | Protein | Peptidesequence | Site | Glycan moiety | MS(calcd.), m/z | Retention time/min | |
|---|---|---|---|---|---|---|---|
| Precursor ion | Product ion | ||||||
| 1 | Apo C3 | GWVTDGFSSLK | 598.8 | 244.2, 854.4 | 7.25 | ||
| 2 | Apo C3 | PTSAVAA | 94 | 0_3_0_0 | 613.3 | 204.1 | 7.21 |
| 3 | Apo C3 | PTSAVAA | 94 | 0_3_1_0 | 686.3 | 204.1 | 11.07 |
| 4 | Apo C3 | PTSAVAA | 94 | 1_1_0_1 | 636.8 | 274.1 | 8.18 |
| 5 | Apo C3 | PTSAVAA | 94 | 1_1_0_2 | 782.3 | 274.1 | 8.29 |
| 6 | Apo C3 | PTSAVAA | 94 | 1_1_1_1 | 709.8 | 274.1 | 7.89 |
| 7 | Apo C3 | PTSAVAA | 94 | 1_2_0_2 | 883.9 | 274.1 | 7.84 |
| 8 | Apo C3 | PTSAVAA | 94 | 1_2_1_0 | 665.8 | 366.1 | 11.08 |
| 9 | Apo C3 | PTSAVAA | 94 | 1_3_0_0 | 694.3 | 366.1 | 11.07 |
| 10 | Apo C3 | PTSAVAA | 94 | 1_3_1_1 | 912.9 | 274.1 | 7.86 |
| 11 | Apo C3 | PTSAVAA | 94 | 2_1_1_0 | 645.3 | 366.1 | 11.07 |
| 12 | Apo C3 | PTSAVAA | 94 | 2_2_0_0 | 673.8 | 366.1 | 7.86 |
| 13 | Apo C3 | PTSAVAA | 94 | 2_2_1_2 | 1037.9 | 274.1 | 6.32 |
| 14 | Apo C3 | PTSAVAA | 94 | 2_2_2_0 | 673.8 | 274.1 | 7.03 |
| 15 | Apo C3 | PTSAVAA | 94 | 2_2_3_0 | 892.9 | 366.1 | 10.58 |
| 16 | Apo C3 | PTSAVAA | 94 | 2_3_0_1 | 920.9 | 274.1 | 7.60 |
| No. | 156 N site | 176 N site | 319 O site | 346 O site |
|---|---|---|---|---|
| 1 | 5_4_1_2 | 5_4_0_1 | 1_1_0_1 | 1_1_0_2 |
| 2 | 6_5_0_2 | 6_5_0_1 | 1_1_0_2 | |
| 3 | 6_5_1_0 | 6_5_0_2 | 1_1_1_1 | |
| 4 | 6_5_1_3 | 6_5_0_3 | 1_2_0_1 | |
| 5 | 6_5_1_2 | |||
| 6 | 6_5_1_3 |
Table 4 New glycoforms found in four glycosylation site of A2HSG
| No. | 156 N site | 176 N site | 319 O site | 346 O site |
|---|---|---|---|---|
| 1 | 5_4_1_2 | 5_4_0_1 | 1_1_0_1 | 1_1_0_2 |
| 2 | 6_5_0_2 | 6_5_0_1 | 1_1_0_2 | |
| 3 | 6_5_1_0 | 6_5_0_2 | 1_1_1_1 | |
| 4 | 6_5_1_3 | 6_5_0_3 | 1_2_0_1 | |
| 5 | 6_5_1_2 | |||
| 6 | 6_5_1_3 |
| No. | 94 O site | No. | 94 O site | No. | 94 O site |
|---|---|---|---|---|---|
| 1 | 0_3_1_0 | 4 | 1_2_1_0 | 7 | 2_2_0_0 |
| 2 | 1_1_1_1 | 5 | 1_3_0_0 | 8 | 2_2_2_0 |
| 3 | 1_2_0_2 | 6 | 2_1_1_0 | 9 | 2_2_3_0 |
Table 5 New glycoforms found in 94 O-glycosylation site of Apo C3
| No. | 94 O site | No. | 94 O site | No. | 94 O site |
|---|---|---|---|---|---|
| 1 | 0_3_1_0 | 4 | 1_2_1_0 | 7 | 2_2_0_0 |
| 2 | 1_1_1_1 | 5 | 1_3_0_0 | 8 | 2_2_2_0 |
| 3 | 1_2_0_2 | 6 | 2_1_1_0 | 9 | 2_2_3_0 |
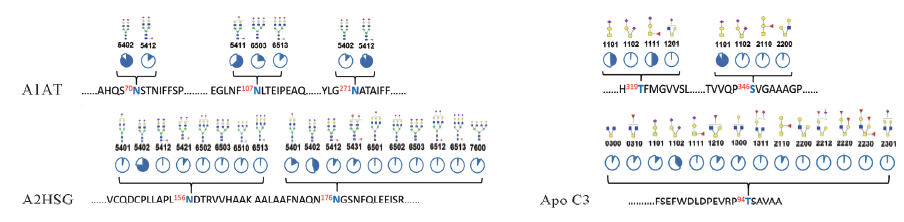
Fig.6 Protein backbone and structures for N- and O-glycopeptides including their site occupancies The glycan monosaccharides annotation includes N-acetylglucosamine(HexNAc)( ), mannose(), galactose(), fucose(), N-acetyl neuraminic acid(). The four number glycan code represents the number of hexoses, HexNAc, fucose, and N-acetyl neuraminic acid residues in that order.
| [1] | Zhu X., Parks J. S., Annu. Rev.Nutr.,2012, 32, 161-182 |
| [2] | Feingold K. R., Grunfeld C., J. Lipid Res.,2011, 52, 1-3 |
| [3] | Toth P. P., Barter P. J., Rosenson R. S., Boden W. E., Chapman M. J., Cuchel M., Karas R. H., J. Clin.Lipidology,2013, 7, 484-525 |
| [4] | Levels J. H. M., Marquart J. A., Abraham P. R.,van den Ende A. E.,Molhuizen H. O. F.,van Deventer S. J. H.,Meijers J. C. M., Infect Immun., 2005, 73, 2321-2326 |
| [5] | De Nardo D., Labzin L. I., Kono H., Seki R., Schmidt S. V., Beyer M., Vogelhuber J., Nature Immun.,2014, 15, 152-160 |
| [6] | Umemoto T., Han C. Y., Mitra P., Averill M. M., Tang C., Goodspeed L., Wei H., Circul. Res.,2013, 112, 1345-1354 |
| [7] | Eklund K. K., Niemi K., Kovanen P. T., Crit. Rev.Immun.,2012, 32, 254-269 |
| [8] | HDL Cholesterol—Dr. Milton Alvis Jr's Insights on Health Tap.() |
| [9] | Huang J., Lee H., Zivkovic A. M., Smilowitz J. T., Rivera N., German J. B., Lebrilla C. B., J. Proteome Res.,2014, 13, 681-691 |
| [10] | Kontush A., Chapman M., J. Pharmacol.Rev.,2006, 58, 342-374 |
| [11] | Krishnan S., Shimoda M., Sacchi R., Kailemia M. J., Luxardi G., Kaysen G. A., Grimes B., Scientific Reports,2017, 7, 43728 |
| [12] | Moran G., Carcamo C., Concha M., Folch H., Revistaiberoamericana de Micologia,2015, 32, 25-29 |
| [13] | O’neill L., Rooney P., Molloy D., Connolly M., McCormick J., McCarthy G., Molloy E., Arthritis & Rheumatology,2015, 67, 2447-2456 |
| [14] | Guttman O., Freixo-Lima G. S., Kaner Z., Lior Y., Rider P., Lewis E. C., Frontiers in Immun.,2016, 7, 1-14 |
| [15] | Häusler M., Schäfer C., Osterwinter C., Jahnen-Dechent W., Pediatric Res.,2009, 66, 660-664 |
| [16] | Cui F., Li K., Li Y., Zhang X., An C., Lipids in Health and Disease,2014, 13, 170 |
| [17] | Luo H. T., Huang X. L., Wu H. Q., Zhu Z. X., Huang F., Lin X. S., Chinese J. Anal. Chem.,2012, 40(2), 273-279 |
| (罗辉泰,黄晓兰,吴惠勤,朱志鑫,黄芳,林晓珊. 分析化学, 2012, 40(2), 273-279) | |
| [18] | Bi Y. F., Zhu H. B., Pi Z. F., Liu Z. Q., Song F. R., Chem. J. Chinese Universities,2013, 34(5), 1067-1071 |
| (毕云枫,朱洪彬,皮子凤,刘志强,宋凤瑞. 高等学校化学学报, 2013, 34(5), 1067-1071) | |
| [19] | Yu L. L., Wen C., Li X., Fang S. Q., Yang L. C., Wang T., Hu K. F., Anal. Bioanal. Chem.,2018, 410(7), 2011-2018 |
| [20] | Idowu I., Francisco O., Thomas P. J., Johnson W., Marvin C., Stetefeld J., Tomy G. T., Rapid Commun. Mass Spectrom., 2018, 32(3), 277-287 |
| [21] | Mei M., Bissada K. K., Malloy T. B., Darnell L. M., Szymcyk E. B., Org. Geochem.,2018, 116, 35-50 |
| [22] | Kailemia M. J., Wei W. H., Nguyen K., Beals E., Sawrey-Kubicek L., Rhodes C., Zhu C. H., Sacchi R., Zivkovic A. M., Lebrilla C. B., J. Proteome Res.,2018, 17(2), 834-845 |
| [23] | Su P., Chen X. N., He Z. J., Yang Y., Chem. Res. Chinese Universities,2017, 33(6), 876-881 |
| [24] | Wei W. H., Chu Y. Q., Wang R. Z., He X. D., Ding C. F., Rapid Commun. Mass Spectrom.,2015, 29, 927-936 |
| [25] | De Leoz M. L. A.,Gaerlan S. C., Strum J. S., Dimapasoc L. M., Mirmiran M., Tancredi D. J., Lebrilla C. B., J. Proteome Res.,2012, 11, 4662-4679 |
| [26] | Sun D., Liu X. Y., Xu S. H., Tian Y., Xu W. Q., Tao Y. C., Chem. Res. Chinese Universities,2018, 34(6), 899-904 |
| [27] | McGarrah R. W., Kelly J. P., Craig D. M., Haynes C., Kraus W. E., Shah S. H., Clin.Chem.,2017, 63, 288-296 |
| [28] | Chen Y. H., Yan G. Q., Zhou X. W., Yang P. Y., Chinese J. Chromatography,2010, 28(2), 135-139 |
| (陈瑶函,晏国全,周新文, 杨芃原. 色谱, 2010, 28(2), 135-139) |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||