

Chem. J. Chinese Universities ›› 2024, Vol. 45 ›› Issue (11): 20240344.doi: 10.7503/cjcu20240344
• Article • Previous Articles Next Articles
XU Xia1, QIN Weida1, LI Ruomeng1, WANG Qianqian2, LIU Ning2, LI Gongyu1( )
)
Received:2024-07-08
Online:2024-11-10
Published:2024-08-23
Contact:
LI Gongyu
E-mail:ligongyu@nankai.edu.cn
Supported by:CLC Number:
TrendMD:
XU Xia, QIN Weida, LI Ruomeng, WANG Qianqian, LIU Ning, LI Gongyu. Mass Spectrometry-based Deep Coverage Proteome: Evaluation of Cellular Protein Extraction Methods[J]. Chem. J. Chinese Universities, 2024, 45(11): 20240344.
| Abb. | Full name | Structure | Lysis buffer | Clean⁃up method | Time |
|---|---|---|---|---|---|
| Urea(U) | Urea⁃based Protein Extraction | 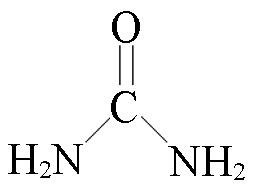 | 50 mmol/L Tris buffer, 8 mol/L urea, 5 mmol/L CaCl2, 30 mmol/L NaCl | C18 SepPak desalting | ca. 10 h |
| SDS(S) | Surfactant and Chaotropic Agent Assisted Sequential Extraction/On⁃Pellet Digestion | 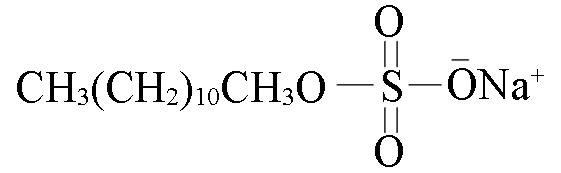 | 50 mmol/L Tris buffer, 4% SDS | 80% Acetone precipitation+C18 SepPak desalting | ca. 22 h |
| Trizol(T) | Trizol, Total RNA Extractor | 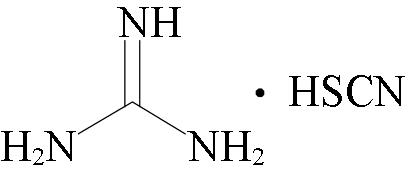 | Trizol, chloroform | Evaporation+C18 SepPak desalting | ca. 13 h |
| BT(B) | BT Surfactant | 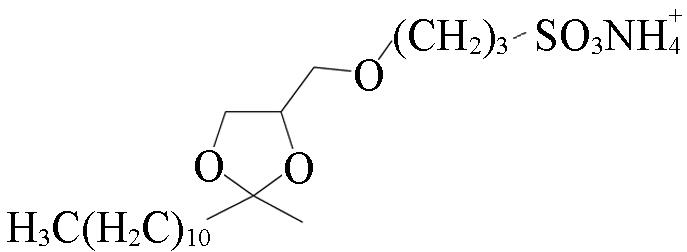 | 50 mmol/L NH4HCO3, 3% BT | pH=2.3—3.0, 45 ℃, On⁃filter degradation, desalting⁃free | ca. 1 h |
Table 1 Information for four protein extraction methods
| Abb. | Full name | Structure | Lysis buffer | Clean⁃up method | Time |
|---|---|---|---|---|---|
| Urea(U) | Urea⁃based Protein Extraction |  | 50 mmol/L Tris buffer, 8 mol/L urea, 5 mmol/L CaCl2, 30 mmol/L NaCl | C18 SepPak desalting | ca. 10 h |
| SDS(S) | Surfactant and Chaotropic Agent Assisted Sequential Extraction/On⁃Pellet Digestion | 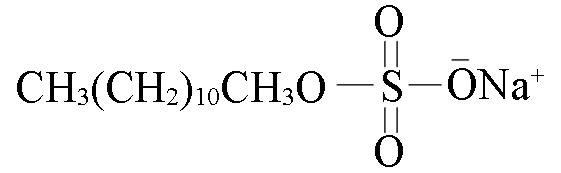 | 50 mmol/L Tris buffer, 4% SDS | 80% Acetone precipitation+C18 SepPak desalting | ca. 22 h |
| Trizol(T) | Trizol, Total RNA Extractor |  | Trizol, chloroform | Evaporation+C18 SepPak desalting | ca. 13 h |
| BT(B) | BT Surfactant |  | 50 mmol/L NH4HCO3, 3% BT | pH=2.3—3.0, 45 ℃, On⁃filter degradation, desalting⁃free | ca. 1 h |
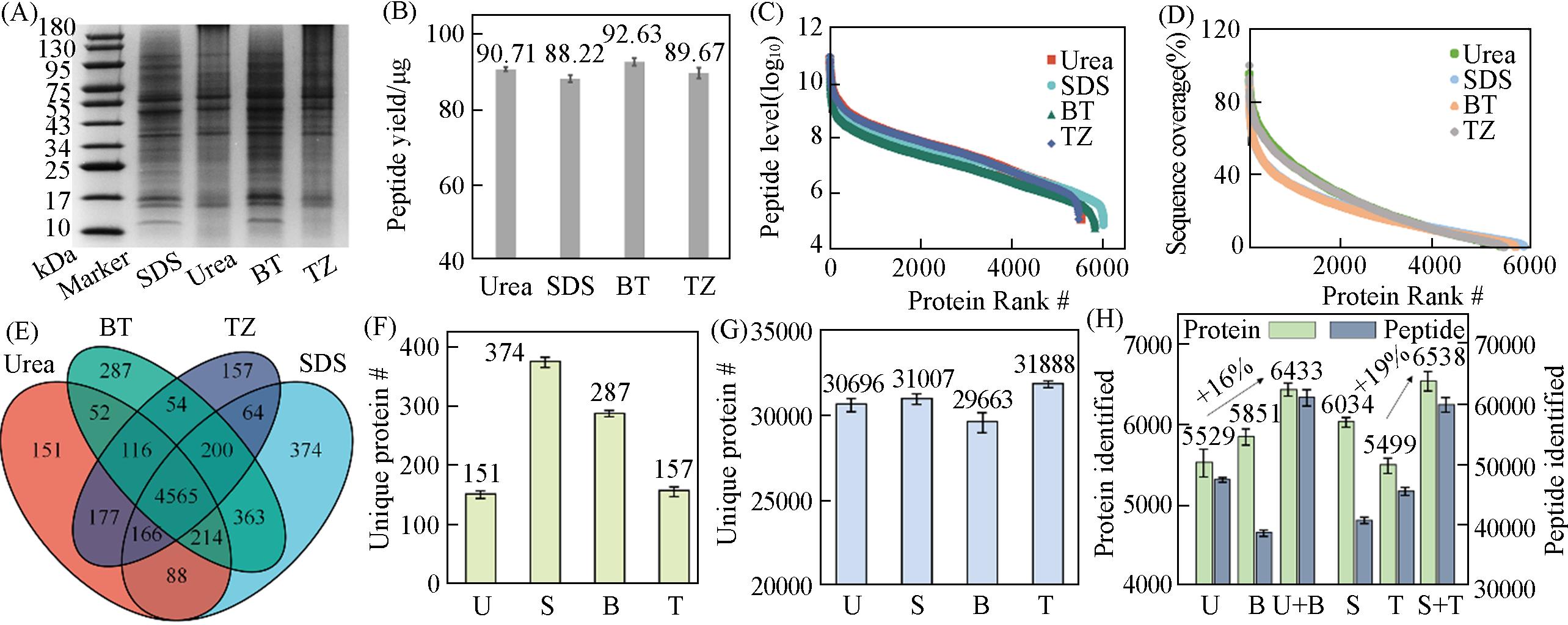
Fig.2 Performance checking on different protein extraction methods, Urea(U), SDS(S), BT(B) and Trizol(T)(A) Representative SDS-PAGE characterization of extracted proteins; (B) peptide yield from the same amount of starting materials; (C) protein abundance distribution; (D) protein sequence coverage; (E) Venn diagram for protein IDs; (F) unique protein ID; (G) unique peptide ID and (H) BT- and Trizol-enhanced protein identification. All data are from triplicate biological replicates with error bars showing standard deviation.
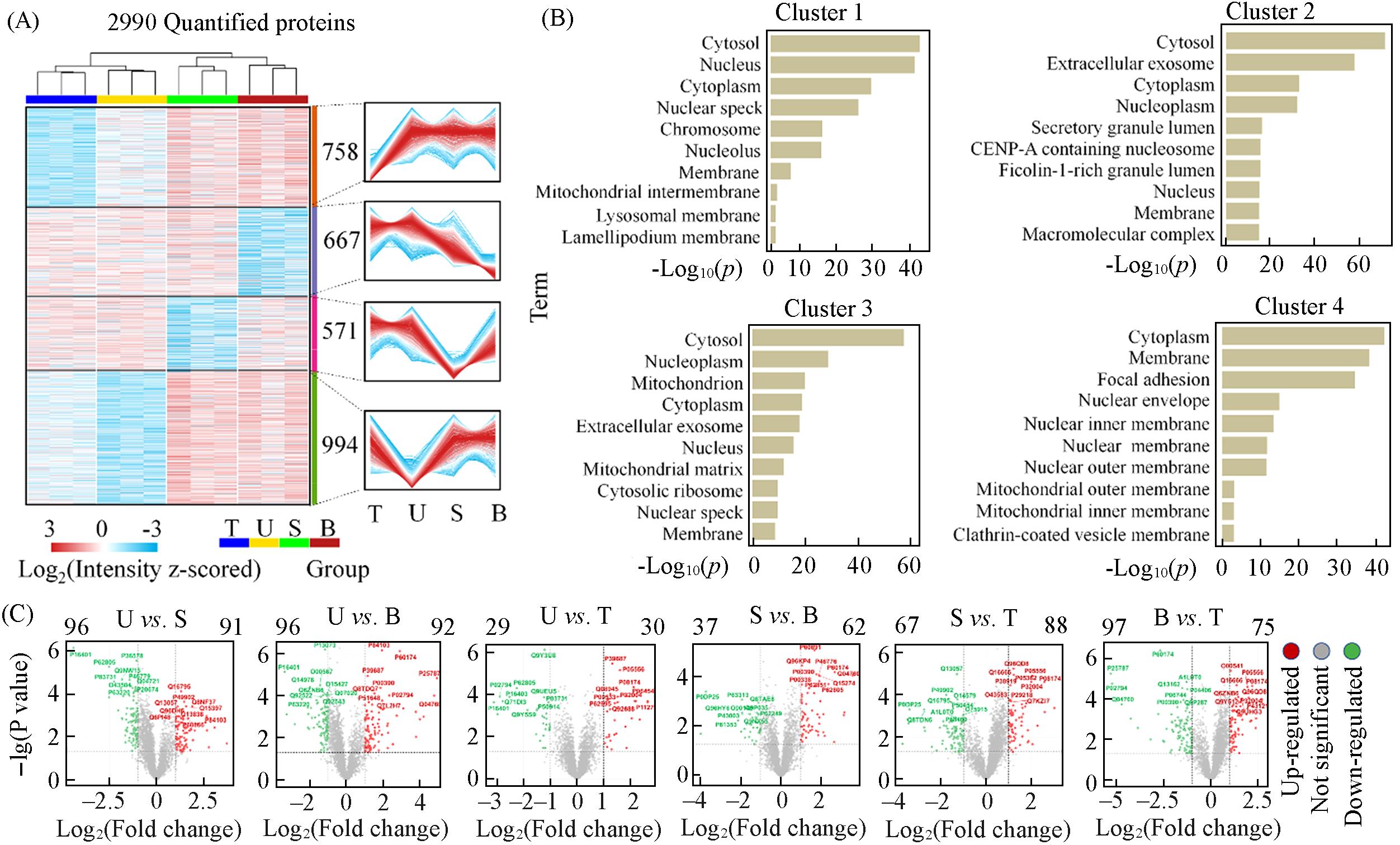
Fig.3 Protein profiles with different extraction methods(A) Hierarchical clustering of 2990 quantified proteins and four clusters of these proteins are depicted on different intensity profiles; (B) the number of proteins and cellular components in each cluster was marked; (C) the volcano plot illustrates the pairwise comparison of protein expression levels between different methods. Dots above the horizontal dashed line represent significantly altered proteins(two-sided t-test, P value<0.05, adjusted for multiple comparisons using the Benjamini-Hochberg method). Downregulated proteins, indicated by green color, while upregulated proteins are represented by red color(protein fold change>2).
| 1 | Shen B., Yi X., Sun Y. T., Bi X. J., Du J. P., Zhang C., Quan S., Zhang F. F., Sun R., Qian L. J., Ge W. G., Liu W., Liang S., Chen H., Zhang Y., Li J., Xu J. Q., He Z. B., Chen B. F., Wang J., Yan H. X., Zheng Y. F., Wang D. L., Zhu J. S., Kong Z. Q., Kang, Z. Y., Liang, X., Ding, X., Ruan G., Xiang N., Cai X., Gao H. H., Li L., Li S. N., Xiao Q., Lu T., Zhu Y., Liu H. F., Chen H. X., Guo T. N., Cell, 2020, 182(1), 59—72 |
| 2 | Li Z. H., Tremmel D. M., Ma F. F., Yu Q. Y., Ma M., Delafield D. G., Shi Y. T., Wang B., Mitchell S. A., Feeney A. K., Jain V. S., Sackett S. D., Odorico J. S., Li L. J., Nat. Commun., 2021, 12(1), 1020 |
| 3 | Martinez⁃Val A., Fort K., Koenig C., Van der Hoeven L., Franciosa G., Moehring T., Ishihama Y., Chen Y. J., Makarov A., Xuan Y., Olsen J. V., Nat. Commun., 2023, 14(1), 3599 |
| 4 | Aebersold R., Mann M., Nature, 2003, 422(6928), 198—207 |
| 5 | Andersen J. S., Wilkinson C. J., Mayor T., Mortensen P., Nigg E. A., Mann M., Nature, 2003, 426(6966), 570—574 |
| 6 | Yang L., Weng S., Qian X. H., Wang M. C., Ying W. T., Anal. Chem., 2022, 94(25), 8827—8832 |
| 7 | Yu Q., Liu X. Y., Keller M. P., Navarrete⁃Perea J., Zhang T., Fu S. P., Vaites L. P., Shuken S. R., Schmid E., Keele G. R., Li J., Huttlin E. L., Rashan E. H., Simcox J., Churchill G. A., Schweppe D. K., Attie A. D., Paulo J. A., Gygi S. P., Nat. Commun., 2023, 14(1), 555 |
| 8 | Meng Y. Q., Chen J. Y., Liu Y. Q., Zhu Y. P., Wong Y. K., Lyu H. N., Shi Q. L., Xia F., Gu L. W., Zhang X. W., Gao P., Tang H., Guo Q. Y., Qiu C., Xu C. C., He X., Zhang J. Z., Wang J. G., J. Pharm. Anal., 2022, 12(6), 879—888 |
| 9 | Davis S., Charles P. D., He L., Mowlds P., Kessler B. M., Fischer R., J. Proteome Res., 2017, 16(3), 1288—1299 |
| 10 | Wang W., Tai F. J., Chen S. N., J. Sep. Sci., 2008, 31(11), 2032—2039 |
| 11 | Doellinger J., Schneider A., Hoeller M., Lasch P., Mol. Cell. Proteomics, 2020, 19(1), 209—222 |
| 12 | Zhang Y., Cai Q. H., Luo Y. X., Zhang Y., Li H. L., J. Pharm. Anal., 2023, 13(1), 63—72 |
| 13 | Wiśniewski J. R., Zougman A., Nagaraj N., Mann M., Nat. Methods, 2009, 6(5), 359—362 |
| 14 | Washburn M. P., Wolters D., Yates J. R. 3rd, Nat. Biotechnol., 2001, 19(3), 242—247 |
| 15 | Ma F. F., Liu F. B., Xu W., Li L. J., J. Proteome Res., 2018, 17(8), 2744—2754 |
| 16 | Ma F. F., Tremmel D. M., Li Z. H., Lietz C. B., Sackett S. D., Odorico J. S., Li L. J., J. Proteome Res., 2019, 18(8), 3156—3165 |
| 17 | Wiśniewski J. R., Anal. Chim. Acta, 2019, 1090, 23—30 |
| 18 | Sethi M. K., Downs M., Zaia J., Mol. Omics, 2020, 16(4), 364—376 |
| 19 | Li L., Sun C. J., Sun Y. T., Dong Z., Wu R. X., Sun X. T., Zhang H. B., Jiang W. H., Zhou Y., Cen X. F., Cai S., Xia H. G., Zhu Y., Guo T. N., Piatkevich K. D., Nat. Commun., 2022, 13(1), 7242 |
| 20 | Grzybowski A. T., Shah R. N., Richter W. F., Ruthenburg A. J., Nat. Protoc., 2019, 14(12), 3275—3302 |
| 21 | Serra A., Gallart⁃Palau X., Dutta B., Sze S. K., J. Proteome Res., 2018, 17(7), 2390—2400 |
| 22 | HaileMariam M., Eguez R. V., Singh H., Bekele S., Ameni G., Pieper R., Yu Y. B., J. Proteome Res., 2018, 17(9), 2917—2924 |
| 23 | Hughes C. S., Moggridge S., Müller T., Sorensen P. H., Morin G. B., Krijgsveld J., Nat. Protoc., 2019, 14(1), 68—85 |
| 24 | Yu D. H., Wang Z., Cupp⁃Sutton K. A., Guo Y. T., Kou Q., Smith K., Liu X. W., Wu S., J. Am. Soc. Mass Spectr., 2021, 32(6), 1336—1344 |
| 25 | Dimayacyac-Esleta B. R. T., Tsai C. F., Kitata R. B., Lin P. Y., Choong W. K., Lin T. D., Wang Y. T., Weng S. H., Yang P. C., Arco S. D., Sung T. Y., Chen Y. J., Anal. Chem., 2015, 87(24), 12016—12023 |
| 26 | Chen Y. J., Roumeliotis T. I., Chang Y. H., Chen C. T., Han C. L., Lin M. H., Chen H. W., Chang G. C., Chang Y. L., Wu C. T., Lin M. W., Hsieh M. S., Wang Y. T., Chen Y. R., Jonassen I., Ghavidel F. Z., Lin, Z. S., Lin K. T., Chen C. W., Sheu P. Y., Hung C. T., Huang K. C., Yang H. C., Lin P. Y., Yen T. C., Lin Y. W., Wang J. H., Raghav L., Lin C. Y., Chen Y. S., Wu P. S., Lai C. T., Weng S. H., Su K. Y., Chang W. H., Tsai P. Y., Robles A. I., Rodriguez H., Hsiao Y. J., Chang W. H., Sung T. Y., Chen J. S., Yu S. L., Choudhary J. S., Chen H. Y., Yang P. C., Chen Y. J., Cell, 2020, 182(1), 226—244 |
| 27 | Liu Z., Wang K. Y., Ye M. L., Anal. Chem., 2023, 95(34), 12580—12585 |
| 28 | Dang Y. M., Jiang N., Wang H., Chen X. C., Gao Y., Zhang X. Y., Qin G. X., Li Y. M., Chen R. B., J. Proteome Res., 2020, 19(3), 1298—1309 |
| 29 | Frei A. P., Jeon O. Y., Kilcher S., Moest H., Henning L. M., Jost C., Plückthun A., Mercer J., Aebersold R., Carreira E. M., Wollscheid B., Nat. Biotechnol., 2012, 30(10), 997—1001 |
| 30 | Ji Y. H., Liu M. J., Bachschmid M. M., Costello C. E., Lin C., Anal. Chem., 2015, 87(11), 5500—5504 |
| 31 | Li M., Powell M. J., Razunguzwa T. T., O’Doherty G. A., J. Org. Chem., 2010, 75(18), 6149—6153 |
| 32 | Tyanova S., Temu T., Cox J., Nat. Protoc., 2016, 11(12), 2301—2319 |
| 33 | Huang D. W., Sherman B. T., Lempicki R. A., Nat. Protoc., 2009, 4(1), 44—57 |
| 34 | Brown K. A., Chen B. F., Guardado⁃Alvarez T. M., Lin Z. Q., Hwang L., Ayaz⁃Guner S., Jin S., Ge Y., Nat. Methods, 2019, 16(5), 417—420 |
| [1] | DONG Peiying, LIU Tong, QIN Weijie. A New Method for Large-scale Enrichment and Stepwise Identification of RNA-protein Complexes [J]. Chem. J. Chinese Universities, 2024, 45(11): 20240091. |
| [2] | SHI Qian, LIU Dongmei, FANG Xiaoni, LIU Baohong. High-throughput Analysis of Tyrosinase Activity and Inhibitors Based on Matrix-assisted Laser Desorption/ionization Mass Spectrometry [J]. Chem. J. Chinese Universities, 2024, 45(11): 20240330. |
| [3] | JIANG Yan, CHEN Yanlin, SONG Gaoyu, CHEN Yanyan, BAI Jing, ZHU Yingdi, LI Juan. Bacterial Protein Profiling [J]. Chem. J. Chinese Universities, 2024, 45(11): 20240345. |
| [4] | YAN Yongjie, GAO Wenbo, LU Chenhui, YANG Cheng, XU Shuting. Detection of Coffee Polyphenols in Nanoliter Cerebrospinal Fluid by Microextraction Coupled to Nanoelectrospray Ionization Mass Spectrometry [J]. Chem. J. Chinese Universities, 2024, 45(11): 20240327. |
| [5] | ZHANG Lei, SHEN Huali. Quantitative Accuracy Evaluation of Mass Spectrometry Based Proteomics Methods Commonly Used in Biomarker Research [J]. Chem. J. Chinese Universities, 2024, 45(11): 20240311. |
| [6] | JIN Ying, ZHANG Junjie, ZHANG Yixin, YUAN Yue, HAN Zhenzhen. Research Progress in Exosome Isolation and Proteomics Analysis [J]. Chem. J. Chinese Universities, 2024, 45(11): 20240305. |
| [7] | HU Yuhong, YU Xiangming, SONG Lili, XING Qinghe, ZHOU Feng. Dynamic Proteome Profiling of Neonatal Hair Shaft Using DEEP SEQ Method [J]. Chem. J. Chinese Universities, 2024, 45(11): 20240326. |
| [8] | FAN Zhirui, FANG Qun, YANG Yi. Single Cell Proteomic Analysis by Mass Spectrometry [J]. Chem. J. Chinese Universities, 2024, 45(11): 20240294. |
| [9] | XU Hongmei, WANG Liangchen, MIN Qianhao. Advances in MALDI MS Matrices for the Detection of Small Molecules [J]. Chem. J. Chinese Universities, 2024, 45(11): 20240285. |
| [10] | HUANG Yuying, YU Chengkun, LIU Siqi, REN Yan. Cell Map of Mouse Peripheral Blood Mononuclear Cells with a Label-free Single-cell Proteomics Method [J]. Chem. J. Chinese Universities, 2024, 45(11): 20240355. |
| [11] | LIU Siying, SU Wen, ZHOU Zhongyan, YANG Zhiyu, PEI Huafu, HE Zhiru, WANG Na, YUE Lei. Protein-Small Molecule Interaction Electrospray Ionization Mass Spectrometry Study of the Ubiquitin/Adenosine Triphoshate Couple over Temperature Variation [J]. Chem. J. Chinese Universities, 2024, 45(11): 20240382. |
| [12] | SHEN Fenglin, FENG Zhaoying, FANG Jing, ZHANG Lei, LIU Xiaohui, ZHOU Xinwen. New Technologies for Mass Spectrometry-based Single-cell Resolved Spatial Proteomics Research [J]. Chem. J. Chinese Universities, 2024, 45(11): 20240299. |
| [13] | HUO Zhiyuan, ZHOU Jinping, MA Xiumin, ZHOU Yan, HUANG Lin. Advances in Single-cell Multi-omics Analysis Based on Mass Spectrometry [J]. Chem. J. Chinese Universities, 2024, 45(11): 20240389. |
| [14] | CAO Ting, SHU Weikang, WAN Jingjing. Plasmonic Composites Aid Semi-quantitative Analysis and Identification of Low-molecular-weight Metabolites by Mass Spectrometry [J]. Chem. J. Chinese Universities, 2024, 45(11): 20240325. |
| [15] | ZHANG Yihan, HUA Tianyu, HOU Shijiao, ZHANG Yangyang, YIN Dan, JI Xiangbo, ZHANG Yanhao, PEI Congcong, ZHANG Shusheng. Tip Fe2O3 Nanorods Driven High-performance Mass Spectrometry Analysis for Constructing Metabolic Fingerprint of PM2.5-exposed Mice [J]. Chem. J. Chinese Universities, 2024, 45(11): 20240376. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||